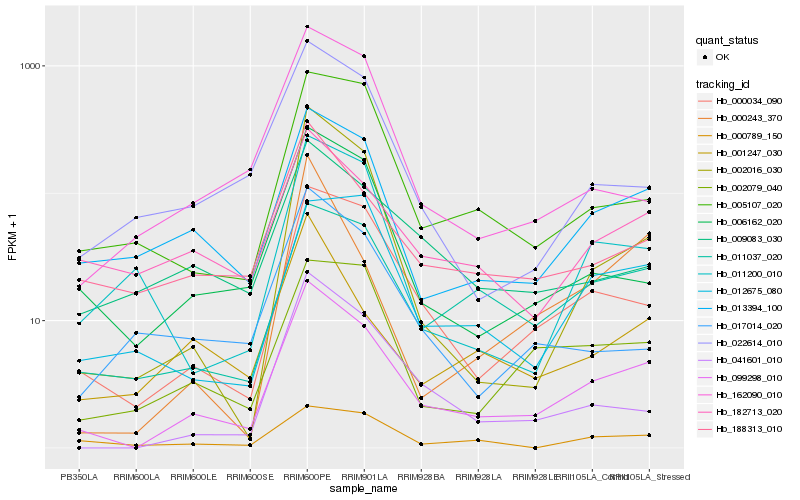
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_099298_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000034_090 |
0.1471660662 |
transcription factor |
TF Family: M-type |
MPF2-like, partial [Dunalia fasciculata] |
| 3 |
Hb_013394_100 |
0.1626040581 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 4 |
Hb_188313_010 |
0.1748377406 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 5 |
Hb_011037_020 |
0.1763873953 |
- |
- |
unknown [Picea sitchensis] |
| 6 |
Hb_006162_020 |
0.1787220374 |
- |
- |
- |
| 7 |
Hb_182713_020 |
0.1883443504 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |
| 8 |
Hb_041601_010 |
0.1904238531 |
- |
- |
- |
| 9 |
Hb_002079_040 |
0.1954369431 |
- |
- |
hypothetical protein VITISV_018542 [Vitis vinifera] |
| 10 |
Hb_005107_020 |
0.1960093357 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 11 |
Hb_002016_030 |
0.1965921772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_022614_010 |
0.1996394633 |
- |
- |
- |
| 13 |
Hb_001247_030 |
0.2001352253 |
- |
- |
- |
| 14 |
Hb_009083_030 |
0.2010098412 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 15 |
Hb_011200_010 |
0.2063034461 |
- |
- |
- |
| 16 |
Hb_012675_080 |
0.2104189393 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 17 |
Hb_162090_010 |
0.2104808391 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |
| 18 |
Hb_000789_150 |
0.2136609469 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000243_370 |
0.2165622817 |
- |
- |
hypothetical protein VITISV_010022 [Vitis vinifera] |
| 20 |
Hb_017014_020 |
0.2178284439 |
- |
- |
- |