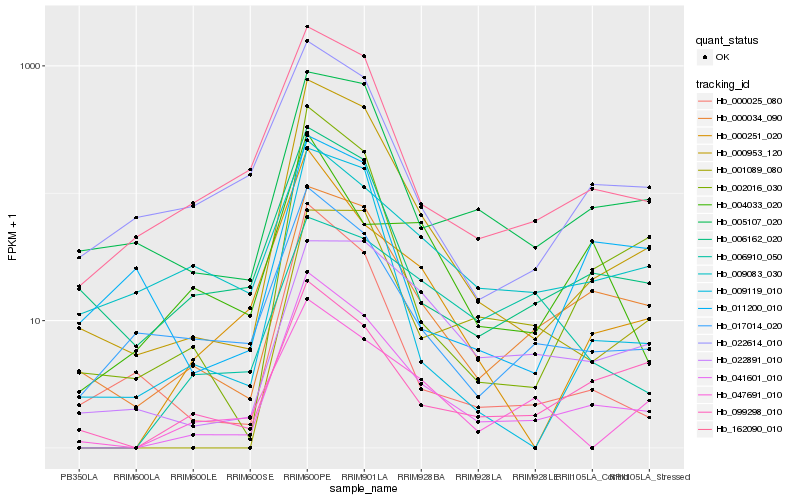
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041601_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000953_120 |
0.1222680292 |
- |
- |
- |
| 3 |
Hb_000251_020 |
0.1609658502 |
- |
- |
hypothetical protein CISIN_1g028541mg [Citrus sinensis] |
| 4 |
Hb_000034_090 |
0.1662405432 |
transcription factor |
TF Family: M-type |
MPF2-like, partial [Dunalia fasciculata] |
| 5 |
Hb_162090_010 |
0.1712195861 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |
| 6 |
Hb_002016_030 |
0.1754256458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000025_080 |
0.1845881387 |
- |
- |
- |
| 8 |
Hb_006162_020 |
0.1867762047 |
- |
- |
- |
| 9 |
Hb_017014_020 |
0.1903223284 |
- |
- |
- |
| 10 |
Hb_099298_010 |
0.1904238531 |
- |
- |
- |
| 11 |
Hb_047691_010 |
0.2048155181 |
- |
- |
Uncharacterized protein F383_01974 [Gossypium arboreum] |
| 12 |
Hb_005107_020 |
0.2050371497 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 13 |
Hb_022614_010 |
0.2052056463 |
- |
- |
- |
| 14 |
Hb_009119_010 |
0.2054276042 |
- |
- |
- |
| 15 |
Hb_001089_080 |
0.2247371306 |
- |
- |
hypothetical protein glysoja_044047 [Glycine soja] |
| 16 |
Hb_009083_030 |
0.2339512687 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 17 |
Hb_022891_010 |
0.2372825936 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 18 |
Hb_004033_020 |
0.2379857027 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 19 |
Hb_011200_010 |
0.2423030152 |
- |
- |
- |
| 20 |
Hb_006910_050 |
0.2441918511 |
- |
- |
hypothetical protein CISIN_1g0480481mg, partial [Citrus sinensis] |