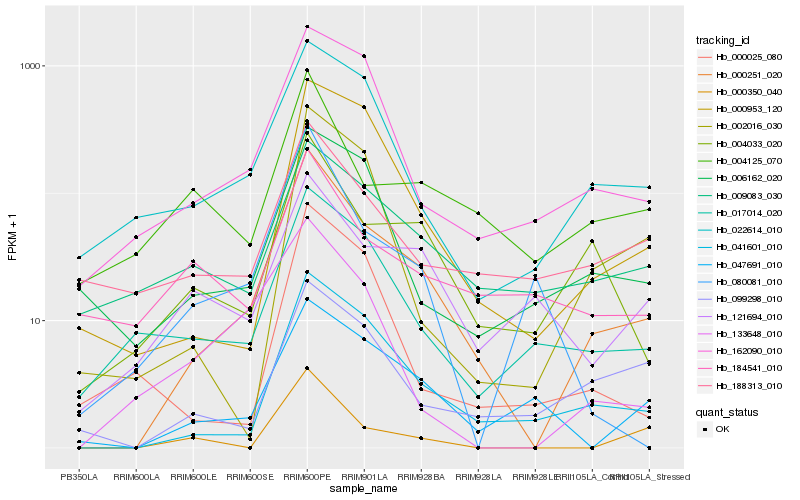
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000251_020 |
0.0 |
- |
- |
hypothetical protein CISIN_1g028541mg [Citrus sinensis] |
| 2 |
Hb_041601_010 |
0.1609658502 |
- |
- |
- |
| 3 |
Hb_000953_120 |
0.2124488135 |
- |
- |
- |
| 4 |
Hb_004033_020 |
0.2154218197 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 5 |
Hb_022614_010 |
0.222679256 |
- |
- |
- |
| 6 |
Hb_162090_010 |
0.2231598896 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |
| 7 |
Hb_017014_020 |
0.2258023396 |
- |
- |
- |
| 8 |
Hb_047691_010 |
0.2335829535 |
- |
- |
Uncharacterized protein F383_01974 [Gossypium arboreum] |
| 9 |
Hb_002016_030 |
0.2372910497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_133648_010 |
0.2416456151 |
- |
- |
- |
| 11 |
Hb_006162_020 |
0.2450139572 |
- |
- |
- |
| 12 |
Hb_099298_010 |
0.2452101011 |
- |
- |
- |
| 13 |
Hb_000025_080 |
0.2466638931 |
- |
- |
- |
| 14 |
Hb_121694_010 |
0.2488822236 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 15 |
Hb_080081_010 |
0.2495967388 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Fragaria vesca subsp. vesca] |
| 16 |
Hb_188313_010 |
0.2522684088 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 17 |
Hb_009083_030 |
0.2544729301 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 18 |
Hb_000350_040 |
0.2568755001 |
- |
- |
- |
| 19 |
Hb_004125_070 |
0.2575232907 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 20 |
Hb_184541_010 |
0.2685666797 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |