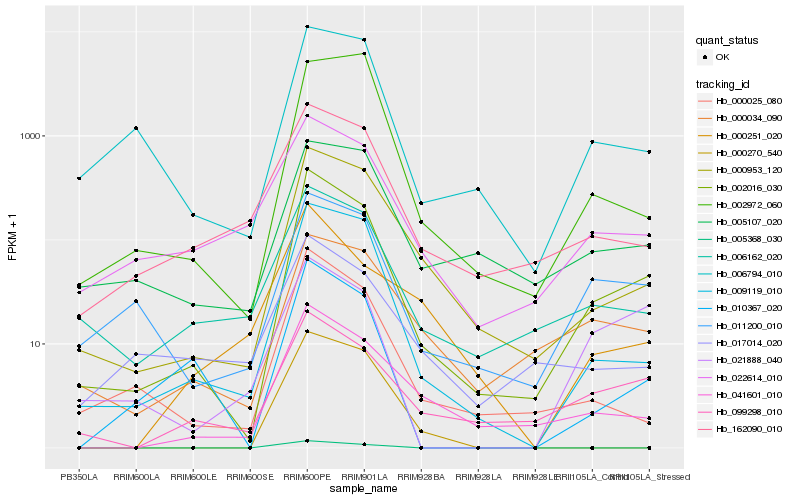
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002016_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000953_120 |
0.1470167549 |
- |
- |
- |
| 3 |
Hb_009119_010 |
0.1542426413 |
- |
- |
- |
| 4 |
Hb_041601_010 |
0.1754256458 |
- |
- |
- |
| 5 |
Hb_000025_080 |
0.1762280363 |
- |
- |
- |
| 6 |
Hb_011200_010 |
0.1821042276 |
- |
- |
- |
| 7 |
Hb_010367_020 |
0.1923490747 |
- |
- |
hypothetical protein POPTR_0009s054801g, partial [Populus trichocarpa] |
| 8 |
Hb_006162_020 |
0.1956437917 |
- |
- |
- |
| 9 |
Hb_099298_010 |
0.1965921772 |
- |
- |
- |
| 10 |
Hb_162090_010 |
0.1979693063 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |
| 11 |
Hb_006794_010 |
0.1982701805 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 12 |
Hb_022614_010 |
0.2019606201 |
- |
- |
- |
| 13 |
Hb_000034_090 |
0.2027961844 |
transcription factor |
TF Family: M-type |
MPF2-like, partial [Dunalia fasciculata] |
| 14 |
Hb_005107_020 |
0.215414274 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 15 |
Hb_002972_060 |
0.225920845 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 16 |
Hb_017014_020 |
0.2298512221 |
- |
- |
- |
| 17 |
Hb_021888_040 |
0.2368262085 |
- |
- |
PREDICTED: uncharacterized protein LOC103499290 isoform X2 [Cucumis melo] |
| 18 |
Hb_000251_020 |
0.2372910497 |
- |
- |
hypothetical protein CISIN_1g028541mg [Citrus sinensis] |
| 19 |
Hb_000270_540 |
0.2471540856 |
- |
- |
hypothetical protein JCGZ_11527 [Jatropha curcas] |
| 20 |
Hb_005368_030 |
0.2480853723 |
- |
- |
- |