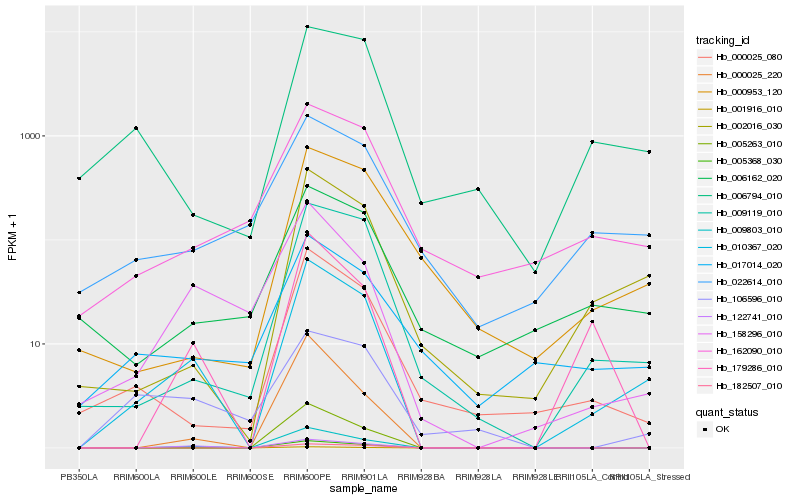
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010367_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s054801g, partial [Populus trichocarpa] |
| 2 |
Hb_009803_010 |
0.18496503 |
- |
- |
hypothetical protein JCGZ_08257 [Jatropha curcas] |
| 3 |
Hb_002016_030 |
0.1923490747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_122741_010 |
0.1996685765 |
- |
- |
hypothetical protein JCGZ_02014 [Jatropha curcas] |
| 5 |
Hb_009119_010 |
0.2008497122 |
- |
- |
- |
| 6 |
Hb_000025_080 |
0.2271517481 |
- |
- |
- |
| 7 |
Hb_158296_010 |
0.2292599207 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 8 |
Hb_179286_010 |
0.2352356347 |
- |
- |
STRUWWELPETER family protein [Populus trichocarpa] |
| 9 |
Hb_001916_010 |
0.242725802 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 10 |
Hb_000025_220 |
0.2437321257 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 11 |
Hb_162090_010 |
0.2442560032 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |
| 12 |
Hb_022614_010 |
0.2466221755 |
- |
- |
- |
| 13 |
Hb_006794_010 |
0.2492780603 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 14 |
Hb_000953_120 |
0.2505280985 |
- |
- |
- |
| 15 |
Hb_005368_030 |
0.2515387045 |
- |
- |
- |
| 16 |
Hb_017014_020 |
0.2532090112 |
- |
- |
- |
| 17 |
Hb_005263_010 |
0.2573143055 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Nicotiana sylvestris] |
| 18 |
Hb_106596_010 |
0.2578009004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006162_020 |
0.2583967784 |
- |
- |
- |
| 20 |
Hb_182507_010 |
0.2601852668 |
- |
- |
- |