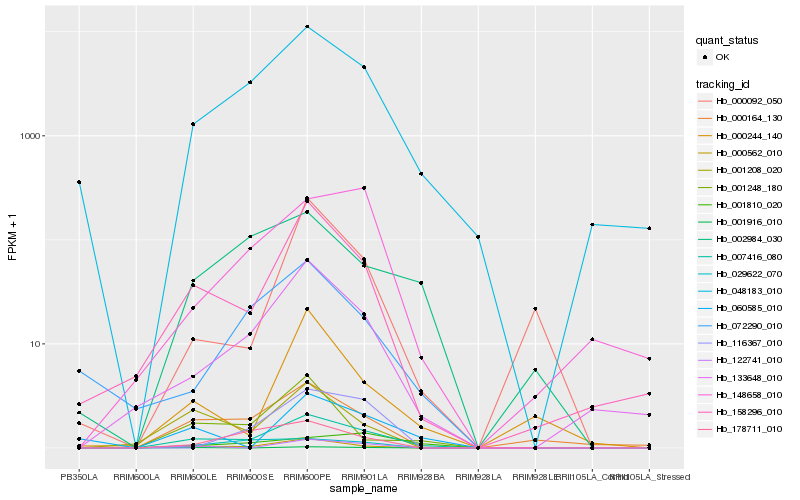
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007416_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_048183_010 |
0.1629791154 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000562_010 |
0.1731325812 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 4 |
Hb_158296_010 |
0.1869368395 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 5 |
Hb_133648_010 |
0.1974471569 |
- |
- |
- |
| 6 |
Hb_000164_130 |
0.2061431104 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 7 |
Hb_001208_020 |
0.227915927 |
- |
- |
- |
| 8 |
Hb_002984_030 |
0.2411020913 |
- |
- |
- |
| 9 |
Hb_178711_010 |
0.2456673087 |
- |
- |
hypothetical protein PHAVU_003G094000g [Phaseolus vulgaris] |
| 10 |
Hb_072290_010 |
0.2562226831 |
- |
- |
PREDICTED: GRF1-interacting factor 1 [Gossypium raimondii] |
| 11 |
Hb_122741_010 |
0.2570982212 |
- |
- |
hypothetical protein JCGZ_02014 [Jatropha curcas] |
| 12 |
Hb_001248_180 |
0.2579770894 |
- |
- |
- |
| 13 |
Hb_029622_070 |
0.2602161607 |
- |
- |
- |
| 14 |
Hb_001916_010 |
0.2684047354 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 15 |
Hb_148658_010 |
0.2781756739 |
- |
- |
- |
| 16 |
Hb_060585_010 |
0.2824556272 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_001810_020 |
0.2827689484 |
- |
- |
- |
| 18 |
Hb_116367_010 |
0.28277658 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Pyrus x bretschneideri] |
| 19 |
Hb_000244_140 |
0.2829012862 |
- |
- |
PREDICTED: cell wall / vacuolar inhibitor of fructosidase 2 [Jatropha curcas] |
| 20 |
Hb_000092_050 |
0.2846795231 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |