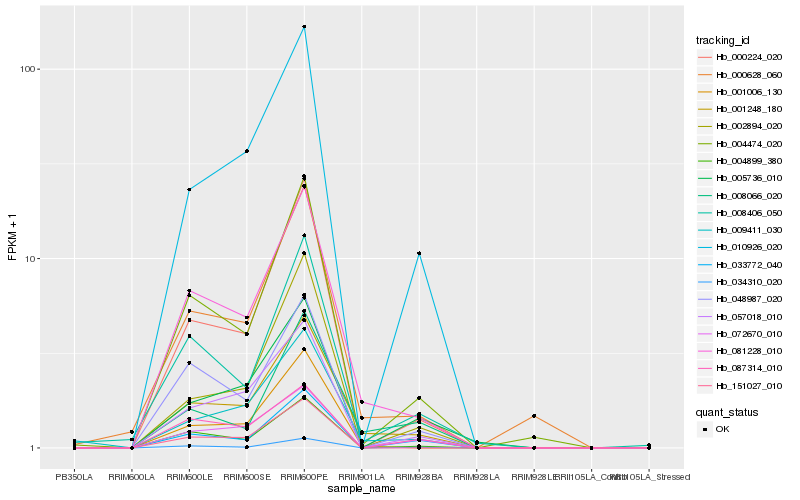
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001248_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_081228_010 |
0.075222794 |
- |
- |
SENESCENCE-RELATED GENE 1 family protein [Populus trichocarpa] |
| 3 |
Hb_009411_030 |
0.1264398645 |
- |
- |
PREDICTED: bifunctional monodehydroascorbate reductase and carbonic anhydrase nectarin-3-like [Malus domestica] |
| 4 |
Hb_000628_060 |
0.1285788652 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 5 |
Hb_008066_020 |
0.1329515585 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004474_020 |
0.1413472273 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 7 |
Hb_010926_020 |
0.1449427595 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Camelina sativa] |
| 8 |
Hb_057018_010 |
0.1491498642 |
- |
- |
hypothetical protein JCGZ_04004 [Jatropha curcas] |
| 9 |
Hb_000224_020 |
0.1496396906 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 10 |
Hb_004899_380 |
0.1526138123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001006_130 |
0.1535238623 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 12 |
Hb_072670_010 |
0.1573614994 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 13 |
Hb_008406_050 |
0.1599229506 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 2 [Jatropha curcas] |
| 14 |
Hb_033772_040 |
0.1619472891 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_048987_020 |
0.1647204351 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 16 |
Hb_034310_020 |
0.1662155825 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 [Jatropha curcas] |
| 17 |
Hb_002894_020 |
0.1675968256 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 18 |
Hb_005736_010 |
0.1680714567 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_087314_010 |
0.1789676943 |
- |
- |
- |
| 20 |
Hb_151027_010 |
0.1803074291 |
- |
- |
(+)-delta-cadinene synthase isozyme A, putative [Ricinus communis] |