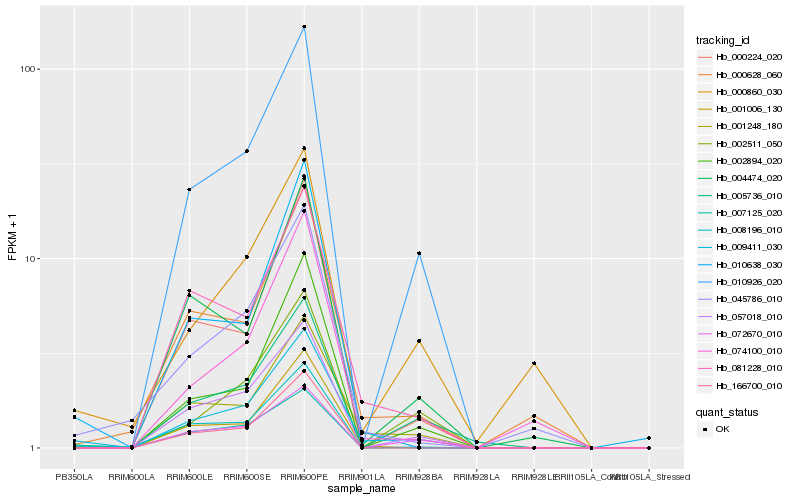
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009411_030 |
0.0 |
- |
- |
PREDICTED: bifunctional monodehydroascorbate reductase and carbonic anhydrase nectarin-3-like [Malus domestica] |
| 2 |
Hb_001248_180 |
0.1264398645 |
- |
- |
- |
| 3 |
Hb_010926_020 |
0.1462713496 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Camelina sativa] |
| 4 |
Hb_057018_010 |
0.1496451884 |
- |
- |
hypothetical protein JCGZ_04004 [Jatropha curcas] |
| 5 |
Hb_008196_010 |
0.1508771164 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_081228_010 |
0.1533371842 |
- |
- |
SENESCENCE-RELATED GENE 1 family protein [Populus trichocarpa] |
| 7 |
Hb_045786_010 |
0.1567689869 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 8 |
Hb_000628_060 |
0.1569529129 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 9 |
Hb_010638_030 |
0.1595848101 |
- |
- |
oleosin1 [Plukenetia volubilis] |
| 10 |
Hb_001006_130 |
0.1647467794 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 11 |
Hb_002894_020 |
0.1680191032 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 12 |
Hb_000224_020 |
0.1692739209 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 13 |
Hb_007125_020 |
0.170132865 |
- |
- |
hypothetical protein POPTR_0003s18010g [Populus trichocarpa] |
| 14 |
Hb_072670_010 |
0.1716623322 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 15 |
Hb_005736_010 |
0.1727425309 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002511_050 |
0.1732337851 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_000860_030 |
0.1738958804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004474_020 |
0.175143148 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 19 |
Hb_074100_010 |
0.1792631345 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 20 |
Hb_166700_010 |
0.1808730388 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |