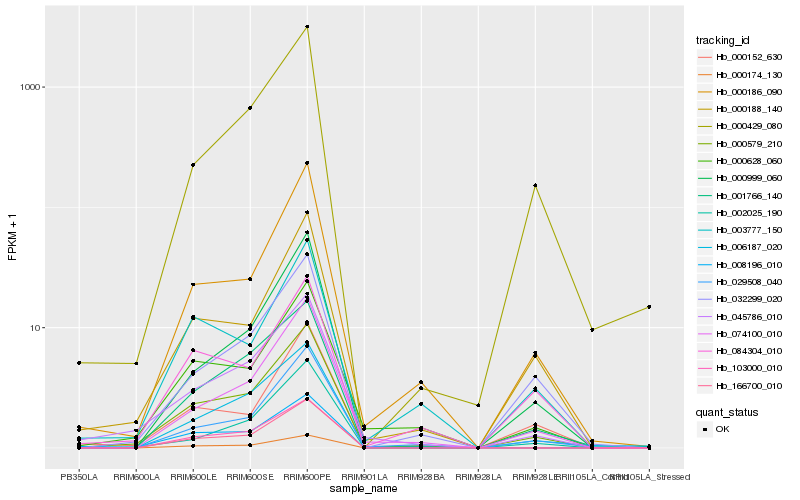
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000579_210 |
0.0 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 2 |
Hb_045786_010 |
0.0897027468 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 3 |
Hb_000188_140 |
0.1008034763 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 4 |
Hb_000186_090 |
0.1017288126 |
- |
- |
ACC oxidase 1 [Hevea brasiliensis] |
| 5 |
Hb_000628_060 |
0.1020519984 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 6 |
Hb_074100_010 |
0.1086587764 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 7 |
Hb_029508_040 |
0.1087885086 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 8 |
Hb_000429_080 |
0.1170703297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001766_140 |
0.1174348919 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 10 |
Hb_032299_020 |
0.1178915032 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 11 |
Hb_008196_010 |
0.1201059363 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_000174_130 |
0.1204015206 |
- |
- |
flowering locus T-like protein [Fagus crenata] |
| 13 |
Hb_166700_010 |
0.1207086253 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 14 |
Hb_006187_020 |
0.1229555047 |
- |
- |
PREDICTED: WAT1-related protein At5g07050-like [Populus euphratica] |
| 15 |
Hb_003777_150 |
0.1235605717 |
- |
- |
PREDICTED: uncharacterized protein LOC105640938 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000152_630 |
0.1251107991 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Jatropha curcas] |
| 17 |
Hb_103000_010 |
0.1257807288 |
- |
- |
hypothetical protein POPTR_0007s02210g [Populus trichocarpa] |
| 18 |
Hb_084304_010 |
0.126226095 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X2 [Jatropha curcas] |
| 19 |
Hb_000999_060 |
0.1263197801 |
- |
- |
hypothetical protein POPTR_0018s11350g [Populus trichocarpa] |
| 20 |
Hb_002025_190 |
0.1263862459 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |