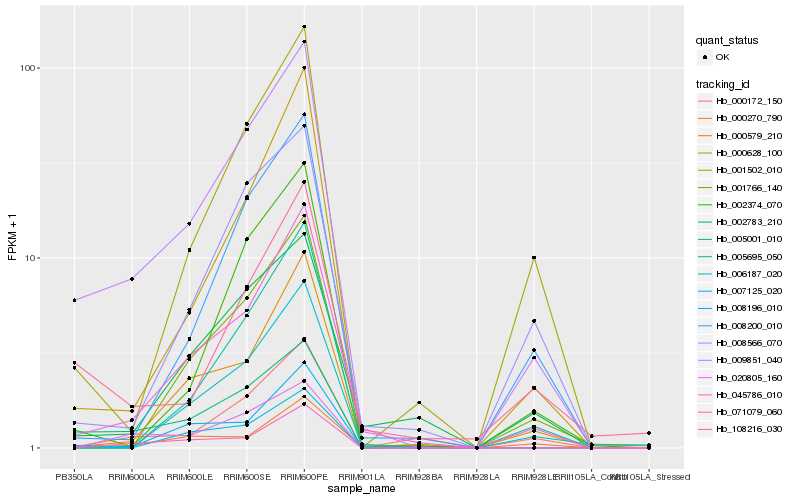
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009851_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005001_010 |
0.108386697 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 3 |
Hb_045786_010 |
0.1153701562 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 4 |
Hb_000628_100 |
0.1643380326 |
- |
- |
hypothetical protein MTR_1g019660 [Medicago truncatula] |
| 5 |
Hb_000172_150 |
0.1689748701 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Populus euphratica] |
| 6 |
Hb_001502_010 |
0.1691764295 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 7 |
Hb_020805_160 |
0.1696180004 |
- |
- |
- |
| 8 |
Hb_000579_210 |
0.1714293924 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 9 |
Hb_002374_070 |
0.1732165918 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_007125_020 |
0.1734274321 |
- |
- |
hypothetical protein POPTR_0003s18010g [Populus trichocarpa] |
| 11 |
Hb_008200_010 |
0.1737328301 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_002783_210 |
0.1742722991 |
- |
- |
PREDICTED: uncharacterized protein LOC105635341 [Jatropha curcas] |
| 13 |
Hb_008566_070 |
0.1757181249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008196_010 |
0.1757327998 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_108216_030 |
0.1765058394 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_001766_140 |
0.1774733522 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 17 |
Hb_071079_060 |
0.1780595119 |
- |
- |
PREDICTED: uncharacterized protein LOC105113464 [Populus euphratica] |
| 18 |
Hb_005695_050 |
0.1823499528 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 19 |
Hb_000270_790 |
0.1828994244 |
- |
- |
hypothetical protein JCGZ_11498 [Jatropha curcas] |
| 20 |
Hb_006187_020 |
0.1843126948 |
- |
- |
PREDICTED: WAT1-related protein At5g07050-like [Populus euphratica] |