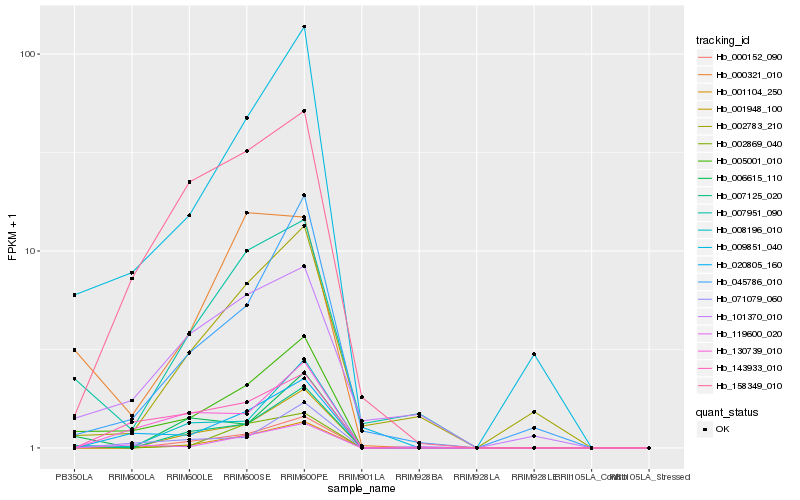
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005001_010 |
0.0 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 2 |
Hb_009851_040 |
0.108386697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_020805_160 |
0.1667287879 |
- |
- |
- |
| 4 |
Hb_007951_090 |
0.1860546737 |
- |
- |
hypothetical protein POPTR_0003s06080g [Populus trichocarpa] |
| 5 |
Hb_007125_020 |
0.1877848188 |
- |
- |
hypothetical protein POPTR_0003s18010g [Populus trichocarpa] |
| 6 |
Hb_045786_010 |
0.194842831 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 7 |
Hb_071079_060 |
0.1962822156 |
- |
- |
PREDICTED: uncharacterized protein LOC105113464 [Populus euphratica] |
| 8 |
Hb_008196_010 |
0.1976051732 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_158349_010 |
0.1996172694 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 10 |
Hb_130739_010 |
0.1998824209 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_000321_010 |
0.2027040603 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_006615_110 |
0.2148870257 |
transcription factor |
TF Family: bZIP |
hypothetical protein JCGZ_11777 [Jatropha curcas] |
| 13 |
Hb_143933_010 |
0.21654773 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_000152_090 |
0.2208657512 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_101370_010 |
0.2229780814 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 16 |
Hb_001948_100 |
0.22526188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_119600_020 |
0.2275467123 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_001104_250 |
0.2277719903 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Ricinus communis] |
| 19 |
Hb_002783_210 |
0.2278687531 |
- |
- |
PREDICTED: uncharacterized protein LOC105635341 [Jatropha curcas] |
| 20 |
Hb_002869_040 |
0.230194092 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |