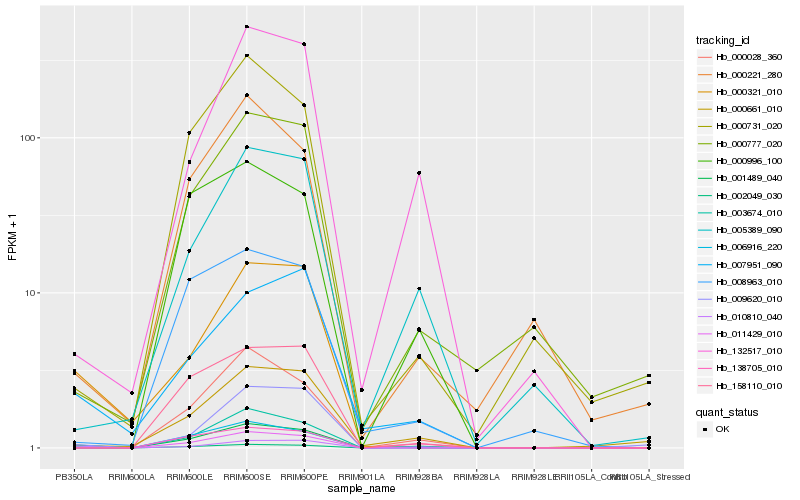
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001489_040 |
0.0 |
- |
- |
PREDICTED: galactose oxidase [Jatropha curcas] |
| 2 |
Hb_003674_010 |
0.1644365041 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 3 |
Hb_000028_360 |
0.1785617861 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_000321_010 |
0.1792364522 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_006916_220 |
0.1828447642 |
- |
- |
hypothetical protein JCGZ_20099 [Jatropha curcas] |
| 6 |
Hb_007951_090 |
0.1844895949 |
- |
- |
hypothetical protein POPTR_0003s06080g [Populus trichocarpa] |
| 7 |
Hb_002049_030 |
0.1846307384 |
- |
- |
PREDICTED: uncharacterized protein LOC105635940 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000661_010 |
0.1869956005 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 9 |
Hb_010810_040 |
0.1871973015 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 10 |
Hb_000777_020 |
0.1904171067 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 11 |
Hb_005389_090 |
0.1904884058 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 12 |
Hb_138705_010 |
0.1907958833 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 13 |
Hb_158110_010 |
0.1912863509 |
- |
- |
hypothetical protein CICLE_v10012017mg [Citrus clementina] |
| 14 |
Hb_000731_020 |
0.1918552023 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 15 |
Hb_000221_280 |
0.1936301628 |
- |
- |
PREDICTED: protein EXORDIUM-like [Populus euphratica] |
| 16 |
Hb_000996_100 |
0.1956428524 |
- |
- |
Nucleotide binding, putative [Theobroma cacao] |
| 17 |
Hb_132517_010 |
0.1958126781 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like isoform X3 [Populus euphratica] |
| 18 |
Hb_011429_010 |
0.1971574142 |
- |
- |
hypothetical protein POPTR_0188s00230g [Populus trichocarpa] |
| 19 |
Hb_009620_010 |
0.1973154778 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_008963_010 |
0.1976345997 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |