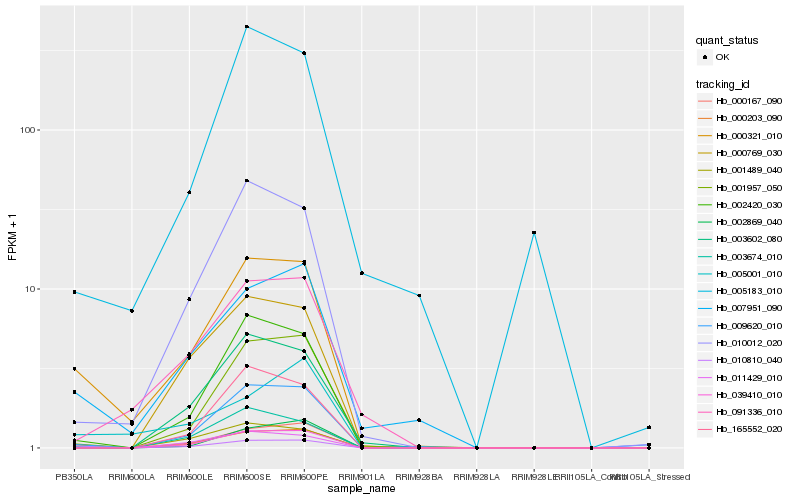
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000321_010 |
0.0 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_010810_040 |
0.1021408679 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 3 |
Hb_007951_090 |
0.1513308447 |
- |
- |
hypothetical protein POPTR_0003s06080g [Populus trichocarpa] |
| 4 |
Hb_010012_020 |
0.1692424415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002420_030 |
0.1712963538 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 6 |
Hb_009620_010 |
0.1734827785 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001489_040 |
0.1792364522 |
- |
- |
PREDICTED: galactose oxidase [Jatropha curcas] |
| 8 |
Hb_003674_010 |
0.1820807869 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 9 |
Hb_001957_050 |
0.1878119719 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 10 |
Hb_091336_010 |
0.1924999155 |
- |
- |
- |
| 11 |
Hb_002869_040 |
0.1989678803 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 12 |
Hb_039410_010 |
0.2012179944 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 13 |
Hb_005001_010 |
0.2027040603 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 14 |
Hb_000203_090 |
0.2062539617 |
- |
- |
hypothetical protein POPTR_0013s04080g [Populus trichocarpa] |
| 15 |
Hb_003602_080 |
0.2127989404 |
- |
- |
hypothetical protein POPTR_0002s10890g [Populus trichocarpa] |
| 16 |
Hb_000167_090 |
0.2162353946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005183_010 |
0.2173740429 |
- |
- |
PREDICTED: probable calcium-binding protein CML17 [Malus domestica] |
| 18 |
Hb_000769_030 |
0.2177518337 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_011429_010 |
0.2194158735 |
- |
- |
hypothetical protein POPTR_0188s00230g [Populus trichocarpa] |
| 20 |
Hb_165552_020 |
0.2207859924 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |