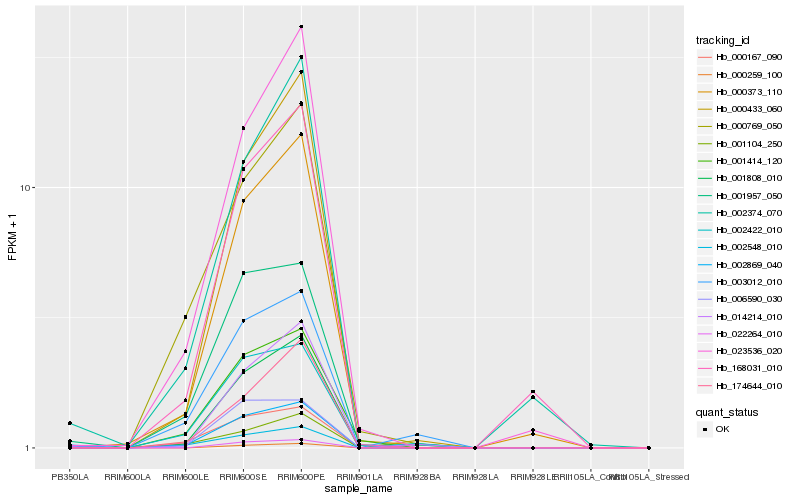
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002869_040 |
0.0 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 2 |
Hb_001957_050 |
0.0909172621 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 3 |
Hb_000167_090 |
0.1244069321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003012_010 |
0.1268857299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002548_010 |
0.131071761 |
- |
- |
hypothetical protein EUGRSUZ_L00530 [Eucalyptus grandis] |
| 6 |
Hb_006590_030 |
0.1321353737 |
- |
- |
unknown [Populus trichocarpa] |
| 7 |
Hb_000373_110 |
0.1330883209 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 8 |
Hb_001808_010 |
0.1356980868 |
- |
- |
PREDICTED: dirigent protein 23-like [Jatropha curcas] |
| 9 |
Hb_002422_010 |
0.1375115671 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 10 |
Hb_001104_250 |
0.1396519725 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Ricinus communis] |
| 11 |
Hb_001414_120 |
0.1410414605 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000433_060 |
0.1418477547 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 13 |
Hb_023536_020 |
0.1447534666 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 14 |
Hb_002374_070 |
0.1463998508 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_000769_050 |
0.1474379457 |
- |
- |
hypothetical protein CISIN_1g030637mg [Citrus sinensis] |
| 16 |
Hb_014214_010 |
0.1476671224 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_168031_010 |
0.1478364626 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6 [Jatropha curcas] |
| 18 |
Hb_022264_010 |
0.1527229989 |
- |
- |
PREDICTED: uncharacterized protein LOC105795741 [Gossypium raimondii] |
| 19 |
Hb_174644_010 |
0.153859897 |
- |
- |
- |
| 20 |
Hb_000259_100 |
0.1539062699 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |