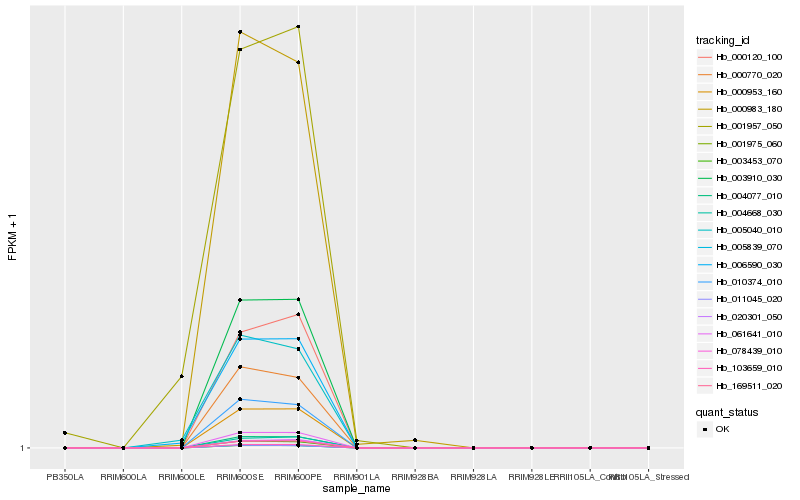
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006590_030 |
0.0 |
- |
- |
unknown [Populus trichocarpa] |
| 2 |
Hb_005040_010 |
0.0477199364 |
- |
- |
hypothetical protein RCOM_1110360 [Ricinus communis] |
| 3 |
Hb_000953_160 |
0.0944393849 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Ricinus communis] |
| 4 |
Hb_003910_030 |
0.0944402833 |
- |
- |
PREDICTED: probable disease resistance protein At5g66910 [Populus euphratica] |
| 5 |
Hb_061641_010 |
0.0944512803 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 6 |
Hb_004077_010 |
0.0945301616 |
- |
- |
retinoblastoma-binding protein, putative [Ricinus communis] |
| 7 |
Hb_169511_020 |
0.094557307 |
- |
- |
PREDICTED: uncharacterized protein LOC105037523, partial [Elaeis guineensis] |
| 8 |
Hb_103659_010 |
0.0945718333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001957_050 |
0.0952166022 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 10 |
Hb_001975_060 |
0.09524319 |
- |
- |
hypothetical protein JCGZ_11079 [Jatropha curcas] |
| 11 |
Hb_003453_070 |
0.0973898697 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor LEP [Jatropha curcas] |
| 12 |
Hb_078439_010 |
0.0982259171 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 13 |
Hb_005839_070 |
0.0984983806 |
- |
- |
hypothetical protein POPTR_0013s08160g [Populus trichocarpa] |
| 14 |
Hb_010374_010 |
0.0986378649 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 15 |
Hb_020301_050 |
0.0991463775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004668_030 |
0.0993081967 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 17 |
Hb_000983_180 |
0.0999475685 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_000770_020 |
0.1010436541 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |
| 19 |
Hb_011045_020 |
0.1014463835 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 20 |
Hb_000120_100 |
0.1015612198 |
- |
- |
- |