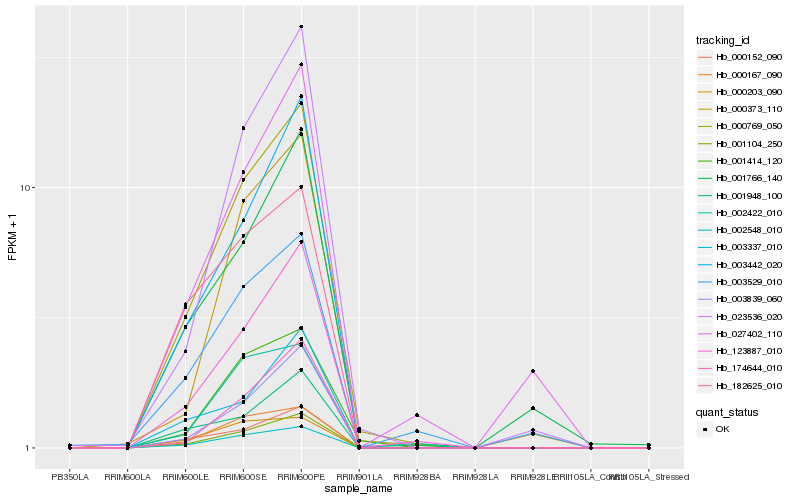
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000769_050 |
0.0 |
- |
- |
hypothetical protein CISIN_1g030637mg [Citrus sinensis] |
| 2 |
Hb_001104_250 |
0.0583389519 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Ricinus communis] |
| 3 |
Hb_002548_010 |
0.0668054872 |
- |
- |
hypothetical protein EUGRSUZ_L00530 [Eucalyptus grandis] |
| 4 |
Hb_123887_010 |
0.089142734 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 5 |
Hb_000152_090 |
0.0936031676 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000167_090 |
0.0976408542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003442_020 |
0.102160894 |
- |
- |
ccd1, putative [Ricinus communis] |
| 8 |
Hb_003839_060 |
0.1035789452 |
- |
- |
PREDICTED: BURP domain-containing protein 11-like [Gossypium raimondii] |
| 9 |
Hb_001414_120 |
0.1072186761 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_023536_020 |
0.1127897078 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 11 |
Hb_001948_100 |
0.1194764735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_174644_010 |
0.1292517206 |
- |
- |
- |
| 13 |
Hb_003337_010 |
0.1309760066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_182625_010 |
0.1320825441 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 15 |
Hb_001766_140 |
0.1322667846 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 16 |
Hb_000373_110 |
0.1331640991 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 17 |
Hb_027402_110 |
0.1376737043 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 18 |
Hb_002422_010 |
0.1392585145 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 19 |
Hb_000203_090 |
0.1393423086 |
- |
- |
hypothetical protein POPTR_0013s04080g [Populus trichocarpa] |
| 20 |
Hb_003529_010 |
0.1402739301 |
- |
- |
PREDICTED: disease resistance protein RPS6-like [Jatropha curcas] |