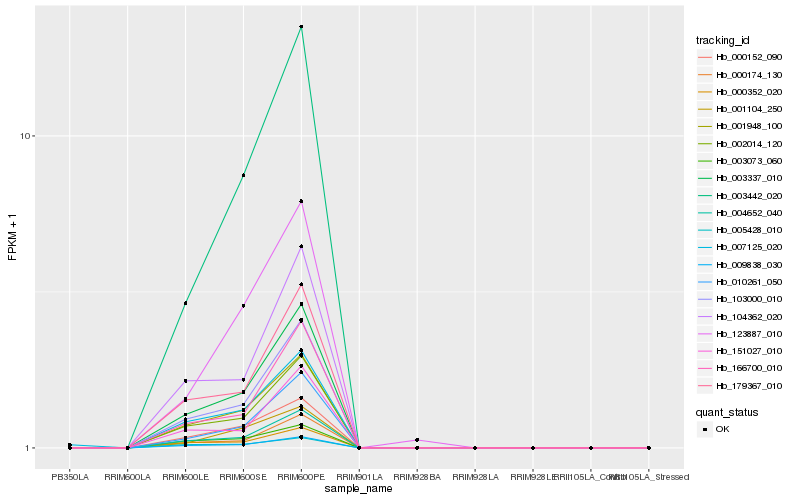
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001948_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009838_030 |
0.0337464424 |
- |
- |
Tetratricopeptide repeat-like superfamily protein [Theobroma cacao] |
| 3 |
Hb_000152_090 |
0.0374762684 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_003337_010 |
0.0393634953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002014_120 |
0.0411813068 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP8 [Populus euphratica] |
| 6 |
Hb_004652_040 |
0.0456153901 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_103000_010 |
0.0523620911 |
- |
- |
hypothetical protein POPTR_0007s02210g [Populus trichocarpa] |
| 8 |
Hb_000352_020 |
0.0568516192 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor LEP [Jatropha curcas] |
| 9 |
Hb_003073_060 |
0.0624550796 |
- |
- |
- |
| 10 |
Hb_179367_010 |
0.0649303204 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 11 |
Hb_005428_010 |
0.0815430625 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_003442_020 |
0.0851623766 |
- |
- |
ccd1, putative [Ricinus communis] |
| 13 |
Hb_104362_020 |
0.0852196176 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_000174_130 |
0.0857858971 |
- |
- |
flowering locus T-like protein [Fagus crenata] |
| 15 |
Hb_007125_020 |
0.0879612331 |
- |
- |
hypothetical protein POPTR_0003s18010g [Populus trichocarpa] |
| 16 |
Hb_010261_050 |
0.0934724067 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_166700_010 |
0.098768843 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 18 |
Hb_001104_250 |
0.104174615 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Ricinus communis] |
| 19 |
Hb_151027_010 |
0.1063541001 |
- |
- |
(+)-delta-cadinene synthase isozyme A, putative [Ricinus communis] |
| 20 |
Hb_123887_010 |
0.1145359619 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |