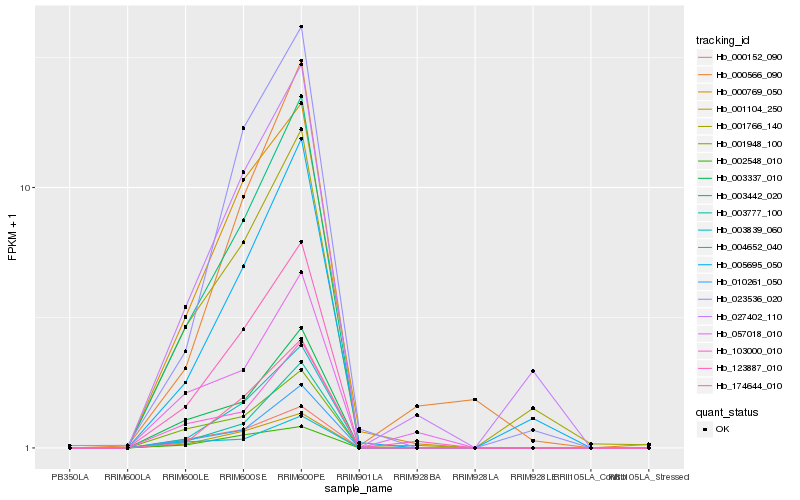
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123887_010 |
0.0 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 2 |
Hb_003442_020 |
0.0709398587 |
- |
- |
ccd1, putative [Ricinus communis] |
| 3 |
Hb_003839_060 |
0.0748818198 |
- |
- |
PREDICTED: BURP domain-containing protein 11-like [Gossypium raimondii] |
| 4 |
Hb_001104_250 |
0.0751359325 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Ricinus communis] |
| 5 |
Hb_000769_050 |
0.089142734 |
- |
- |
hypothetical protein CISIN_1g030637mg [Citrus sinensis] |
| 6 |
Hb_010261_050 |
0.1009435936 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_174644_010 |
0.1054749894 |
- |
- |
- |
| 8 |
Hb_000152_090 |
0.1058888404 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_027402_110 |
0.107096366 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 10 |
Hb_003337_010 |
0.1080518047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001948_100 |
0.1145359619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003777_100 |
0.1148679512 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 13 |
Hb_002548_010 |
0.1150750769 |
- |
- |
hypothetical protein EUGRSUZ_L00530 [Eucalyptus grandis] |
| 14 |
Hb_023536_020 |
0.1165166087 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 15 |
Hb_057018_010 |
0.1188908675 |
- |
- |
hypothetical protein JCGZ_04004 [Jatropha curcas] |
| 16 |
Hb_103000_010 |
0.1200776393 |
- |
- |
hypothetical protein POPTR_0007s02210g [Populus trichocarpa] |
| 17 |
Hb_001766_140 |
0.1224281493 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 18 |
Hb_000566_090 |
0.1239341372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004652_040 |
0.1264333398 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_005695_050 |
0.1273404174 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |