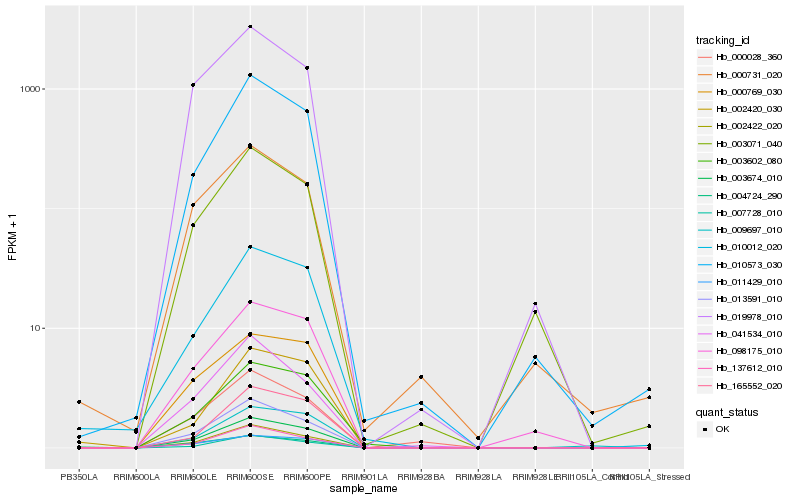
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003674_010 |
0.0 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 2 |
Hb_010012_020 |
0.0858380606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002422_020 |
0.0911808596 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 4 |
Hb_013591_010 |
0.0947199229 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |
| 5 |
Hb_011429_010 |
0.0967059463 |
- |
- |
hypothetical protein POPTR_0188s00230g [Populus trichocarpa] |
| 6 |
Hb_010573_030 |
0.1013865324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002420_030 |
0.1107928957 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 8 |
Hb_004724_290 |
0.1109768689 |
transcription factor |
TF Family: NAC |
- |
| 9 |
Hb_137612_010 |
0.1118242944 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_000769_030 |
0.1150420618 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_003602_080 |
0.1163949232 |
- |
- |
hypothetical protein POPTR_0002s10890g [Populus trichocarpa] |
| 12 |
Hb_098175_010 |
0.1168134939 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 13 |
Hb_007728_010 |
0.118407331 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 14 |
Hb_019978_010 |
0.1220100804 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_165552_020 |
0.1281940164 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 16 |
Hb_000731_020 |
0.1294627022 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 17 |
Hb_000028_360 |
0.1360075665 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_041534_010 |
0.1376173754 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 19 |
Hb_003071_040 |
0.1389321935 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 20 |
Hb_009697_010 |
0.1403564728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |