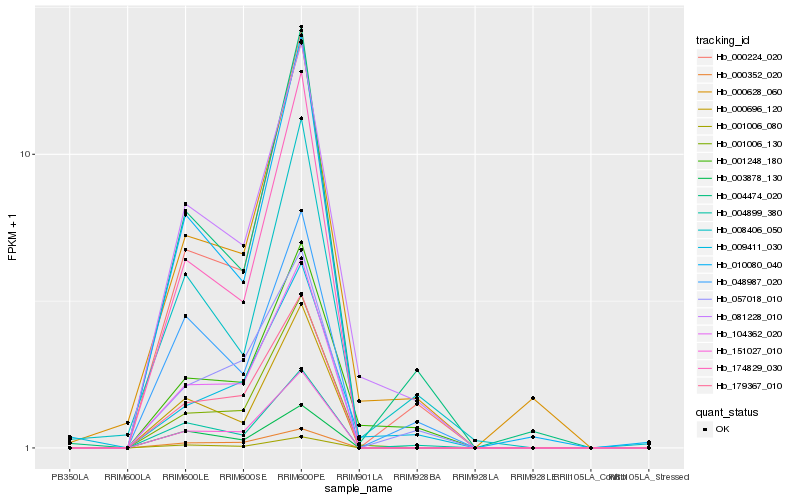
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_081228_010 |
0.0 |
- |
- |
SENESCENCE-RELATED GENE 1 family protein [Populus trichocarpa] |
| 2 |
Hb_001248_180 |
0.075222794 |
- |
- |
- |
| 3 |
Hb_004899_380 |
0.1145613773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000628_060 |
0.1192093389 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 5 |
Hb_004474_020 |
0.1233750291 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 6 |
Hb_048987_020 |
0.1266881174 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 7 |
Hb_001006_130 |
0.1314474724 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 8 |
Hb_000224_020 |
0.1361358698 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 9 |
Hb_001006_080 |
0.1367602681 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 10 |
Hb_104362_020 |
0.14299438 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_008406_050 |
0.1458503614 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 2 [Jatropha curcas] |
| 12 |
Hb_151027_010 |
0.1458965288 |
- |
- |
(+)-delta-cadinene synthase isozyme A, putative [Ricinus communis] |
| 13 |
Hb_174829_030 |
0.1466937228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003878_130 |
0.1467707997 |
- |
- |
- |
| 15 |
Hb_000696_120 |
0.1469146817 |
- |
- |
PREDICTED: protein FD [Populus euphratica] |
| 16 |
Hb_179367_010 |
0.1492155228 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 17 |
Hb_057018_010 |
0.1495133297 |
- |
- |
hypothetical protein JCGZ_04004 [Jatropha curcas] |
| 18 |
Hb_000352_020 |
0.1521999371 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor LEP [Jatropha curcas] |
| 19 |
Hb_010080_040 |
0.1523116993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_009411_030 |
0.1533371842 |
- |
- |
PREDICTED: bifunctional monodehydroascorbate reductase and carbonic anhydrase nectarin-3-like [Malus domestica] |