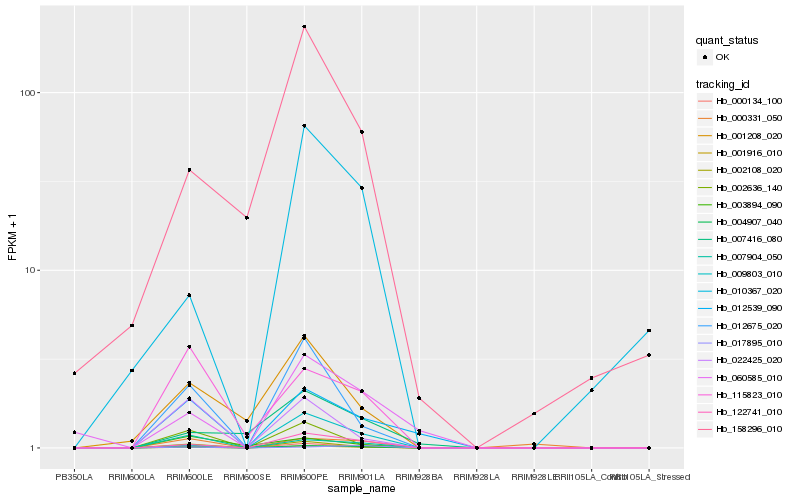
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004907_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s079751g, partial [Populus trichocarpa] |
| 2 |
Hb_001916_010 |
0.0688956312 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 3 |
Hb_122741_010 |
0.1231081629 |
- |
- |
hypothetical protein JCGZ_02014 [Jatropha curcas] |
| 4 |
Hb_000134_100 |
0.1337360092 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Prunus mume] |
| 5 |
Hb_002108_020 |
0.1732558823 |
- |
- |
hypothetical protein JCGZ_16012 [Jatropha curcas] |
| 6 |
Hb_012539_090 |
0.22570006 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: U-box domain-containing protein 26, partial [Eucalyptus grandis] |
| 7 |
Hb_012675_020 |
0.2305719081 |
- |
- |
hypothetical protein CISIN_1g018227mg [Citrus sinensis] |
| 8 |
Hb_007904_050 |
0.2341302064 |
- |
- |
hypothetical protein JCGZ_17883 [Jatropha curcas] |
| 9 |
Hb_115823_010 |
0.2497597856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002636_140 |
0.2510907672 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691-like [Populus euphratica] |
| 11 |
Hb_009803_010 |
0.2554723823 |
- |
- |
hypothetical protein JCGZ_08257 [Jatropha curcas] |
| 12 |
Hb_060585_010 |
0.2555840779 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_001208_020 |
0.2594580084 |
- |
- |
- |
| 14 |
Hb_022425_020 |
0.269016599 |
- |
- |
- |
| 15 |
Hb_010367_020 |
0.2750135309 |
- |
- |
hypothetical protein POPTR_0009s054801g, partial [Populus trichocarpa] |
| 16 |
Hb_158296_010 |
0.2786602523 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 17 |
Hb_000331_050 |
0.2824420016 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 18 |
Hb_007416_080 |
0.2863954151 |
- |
- |
- |
| 19 |
Hb_017895_010 |
0.2960568776 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Prunus mume] |
| 20 |
Hb_003894_090 |
0.2974816403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |