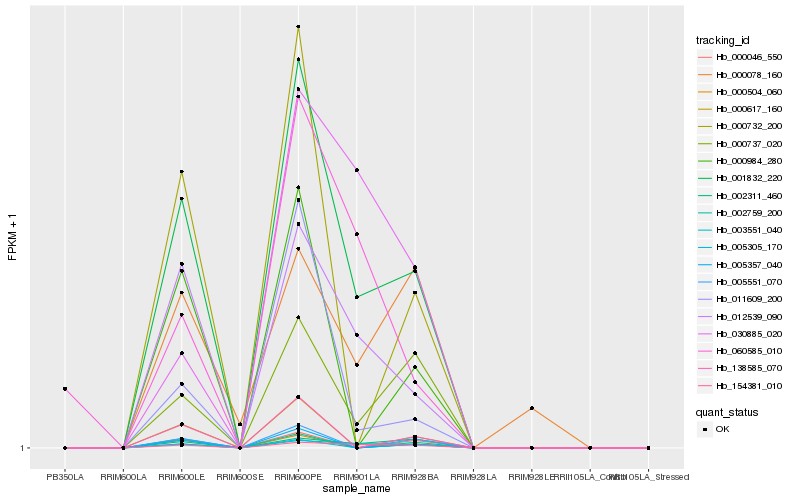
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001832_220 |
0.0 |
- |
- |
- |
| 2 |
Hb_012539_090 |
0.1378318006 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: U-box domain-containing protein 26, partial [Eucalyptus grandis] |
| 3 |
Hb_003551_040 |
0.1381825822 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 4 |
Hb_154381_010 |
0.1451782803 |
- |
- |
- |
| 5 |
Hb_002311_460 |
0.1774259514 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_000737_020 |
0.1803937125 |
- |
- |
- |
| 7 |
Hb_011609_200 |
0.2487852582 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8-like isoform X3 [Populus euphratica] |
| 8 |
Hb_030885_020 |
0.2529366048 |
- |
- |
- |
| 9 |
Hb_000732_200 |
0.254368219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000617_160 |
0.2551168978 |
- |
- |
- |
| 11 |
Hb_138585_070 |
0.2553839563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000504_060 |
0.2558092493 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 13 |
Hb_005551_070 |
0.2558824464 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002759_200 |
0.2564003432 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 15 |
Hb_000078_160 |
0.2572625323 |
- |
- |
PREDICTED: protein LURP-one-related 12-like [Jatropha curcas] |
| 16 |
Hb_000984_280 |
0.2574056636 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 [Jatropha curcas] |
| 17 |
Hb_005305_170 |
0.2577105592 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 8 [Jatropha curcas] |
| 18 |
Hb_005357_040 |
0.2599194209 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 19 |
Hb_060585_010 |
0.2601432572 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_000046_550 |
0.2609981053 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |