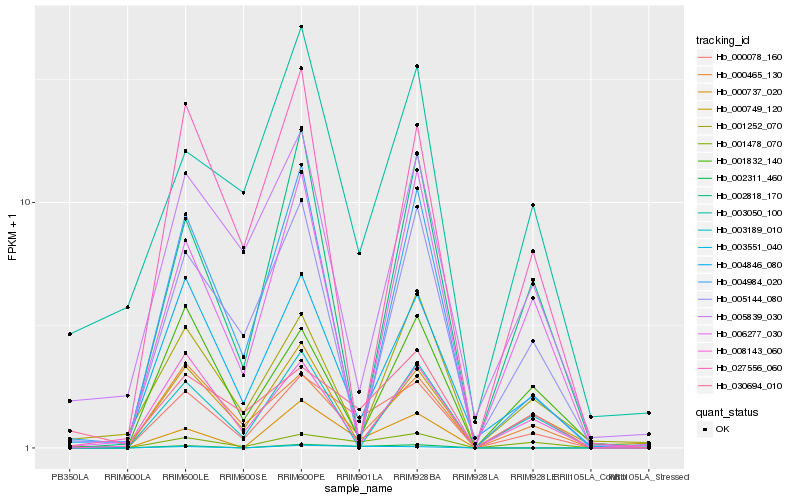
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000078_160 |
0.0 |
- |
- |
PREDICTED: protein LURP-one-related 12-like [Jatropha curcas] |
| 2 |
Hb_001478_070 |
0.154518276 |
- |
- |
PREDICTED: uncharacterized protein LOC105789550 [Gossypium raimondii] |
| 3 |
Hb_000465_130 |
0.1690697983 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0001s15550g [Populus trichocarpa] |
| 4 |
Hb_004846_080 |
0.1904035538 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 5 |
Hb_030694_010 |
0.1979523788 |
- |
- |
PREDICTED: endoglucanase 25-like [Solanum tuberosum] |
| 6 |
Hb_002311_460 |
0.199411733 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_008143_060 |
0.2248520173 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 8 |
Hb_001832_140 |
0.227609476 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |
| 9 |
Hb_000749_120 |
0.2290301876 |
- |
- |
PREDICTED: uncharacterized protein LOC105644255 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000737_020 |
0.2324599561 |
- |
- |
- |
| 11 |
Hb_003050_100 |
0.2428536663 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |
| 12 |
Hb_001252_070 |
0.2431785858 |
- |
- |
hypothetical protein JCGZ_01385 [Jatropha curcas] |
| 13 |
Hb_003189_010 |
0.2440573828 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 14 |
Hb_005144_080 |
0.2463814192 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 15 |
Hb_005839_030 |
0.2476864494 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |
| 16 |
Hb_006277_030 |
0.2480883451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004984_020 |
0.249034045 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 18 |
Hb_003551_040 |
0.2504633167 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 19 |
Hb_002818_170 |
0.2510308668 |
- |
- |
hypothetical protein POPTR_0014s10650g [Populus trichocarpa] |
| 20 |
Hb_027556_060 |
0.2512817749 |
- |
- |
hypothetical protein POPTR_0001s03520g [Populus trichocarpa] |