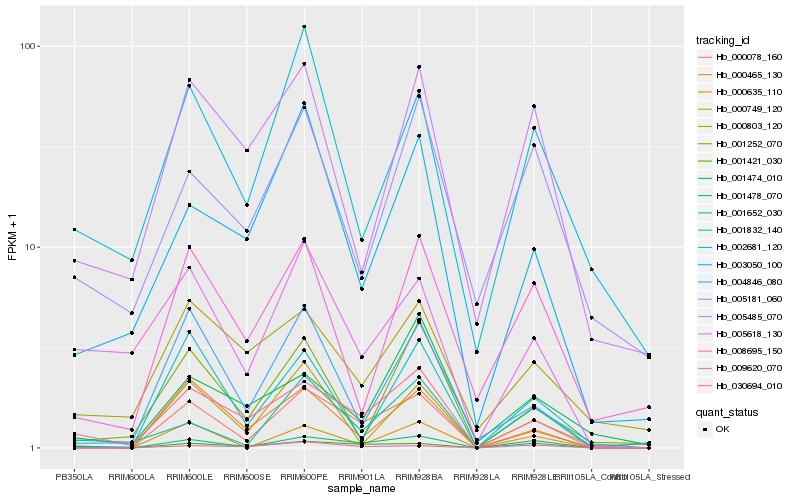
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001478_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105789550 [Gossypium raimondii] |
| 2 |
Hb_030694_010 |
0.137452403 |
- |
- |
PREDICTED: endoglucanase 25-like [Solanum tuberosum] |
| 3 |
Hb_000078_160 |
0.154518276 |
- |
- |
PREDICTED: protein LURP-one-related 12-like [Jatropha curcas] |
| 4 |
Hb_001652_030 |
0.2205043924 |
- |
- |
hypothetical protein CISIN_1g019718mg [Citrus sinensis] |
| 5 |
Hb_001421_030 |
0.2268124613 |
- |
- |
- |
| 6 |
Hb_000465_130 |
0.2277346468 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0001s15550g [Populus trichocarpa] |
| 7 |
Hb_000635_110 |
0.2337501226 |
- |
- |
ELMO domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_005618_130 |
0.234997783 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 9 |
Hb_004846_080 |
0.2379860893 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 10 |
Hb_003050_100 |
0.2398803083 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |
| 11 |
Hb_001832_140 |
0.2469143805 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |
| 12 |
Hb_001252_070 |
0.2494733074 |
- |
- |
hypothetical protein JCGZ_01385 [Jatropha curcas] |
| 13 |
Hb_002681_120 |
0.2506594962 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 14 |
Hb_009620_070 |
0.2508540314 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 15 |
Hb_001474_010 |
0.2533540813 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005485_070 |
0.2537111184 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000803_120 |
0.2553486408 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000749_120 |
0.2568393969 |
- |
- |
PREDICTED: uncharacterized protein LOC105644255 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005181_060 |
0.2596797839 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 20 |
Hb_008695_150 |
0.2605190766 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X1 [Jatropha curcas] |