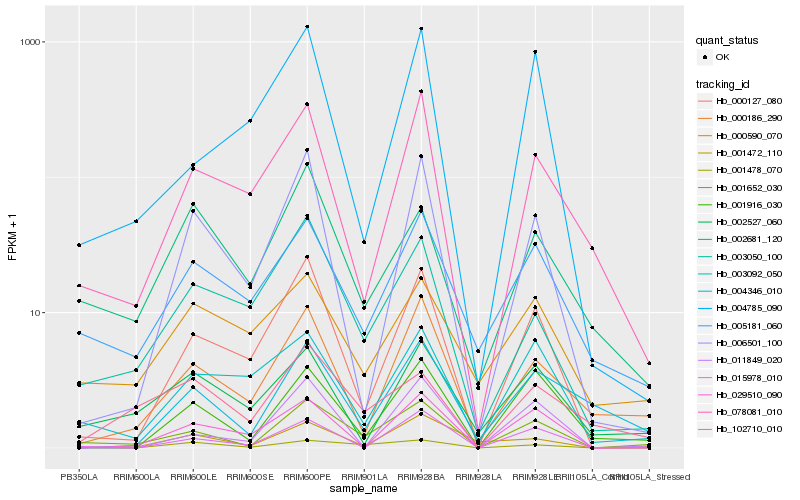
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001652_030 |
0.0 |
- |
- |
hypothetical protein CISIN_1g019718mg [Citrus sinensis] |
| 2 |
Hb_000127_080 |
0.2115978045 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 3 |
Hb_005181_060 |
0.2126495341 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 4 |
Hb_001916_030 |
0.2184435679 |
- |
- |
PREDICTED: uncharacterized protein LOC105802975 [Gossypium raimondii] |
| 5 |
Hb_001478_070 |
0.2205043924 |
- |
- |
PREDICTED: uncharacterized protein LOC105789550 [Gossypium raimondii] |
| 6 |
Hb_000186_290 |
0.220684037 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003050_100 |
0.2210060853 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |
| 8 |
Hb_078081_010 |
0.2290189962 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 9 |
Hb_003092_050 |
0.2328681866 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 10 |
Hb_015978_010 |
0.23322783 |
- |
- |
PREDICTED: probable beta-D-xylosidase 2 [Populus euphratica] |
| 11 |
Hb_006501_100 |
0.235910688 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 12 |
Hb_004785_090 |
0.2406668389 |
- |
- |
hypothetical protein MIMGU_mgv1a018797mg [Erythranthe guttata] |
| 13 |
Hb_001472_110 |
0.2408762178 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 14 |
Hb_004346_010 |
0.2443916516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_029510_090 |
0.2459114691 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 16 |
Hb_002681_120 |
0.2463208689 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 17 |
Hb_011849_020 |
0.2475379881 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 18 |
Hb_102710_010 |
0.247793335 |
- |
- |
hypothetical protein POPTR_0006s22820g [Populus trichocarpa] |
| 19 |
Hb_002527_060 |
0.2506121198 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 20 |
Hb_000590_070 |
0.2527544087 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |