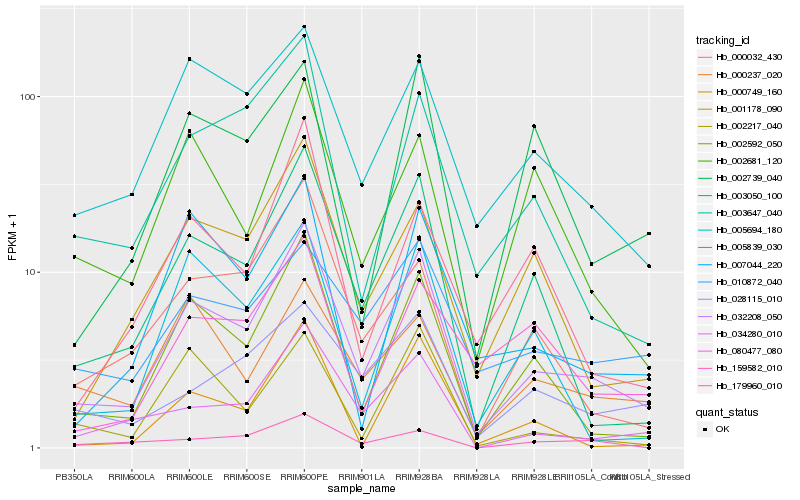
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032208_050 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 2 |
Hb_002592_050 |
0.1306979559 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 3 |
Hb_003050_100 |
0.1348725244 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |
| 4 |
Hb_034280_010 |
0.1489405592 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 5 |
Hb_005694_180 |
0.1621052117 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 6 |
Hb_001178_090 |
0.1670765573 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 7 |
Hb_003647_040 |
0.1744282592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000032_430 |
0.1751161547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007044_220 |
0.1781004008 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_005839_030 |
0.1807486878 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |
| 11 |
Hb_010872_040 |
0.1848561852 |
- |
- |
hypothetical protein F775_31993 [Aegilops tauschii] |
| 12 |
Hb_028115_010 |
0.185464175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002681_120 |
0.1855198847 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 14 |
Hb_002217_040 |
0.1864741972 |
- |
- |
PREDICTED: boron transporter 1-like [Jatropha curcas] |
| 15 |
Hb_002739_040 |
0.1894491251 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 16 |
Hb_000749_160 |
0.1913498852 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 17 |
Hb_000237_020 |
0.1927231333 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 18 |
Hb_179960_010 |
0.1931735995 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 19 |
Hb_080477_080 |
0.1946257685 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 20 |
Hb_159582_010 |
0.1958415233 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |