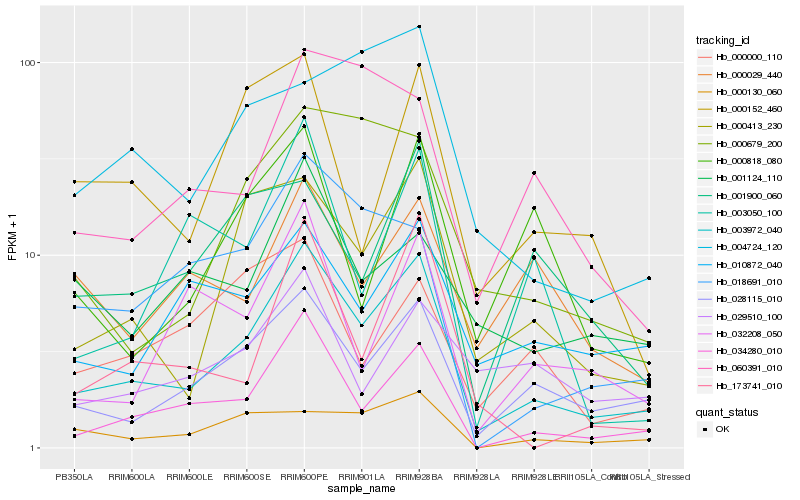
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003972_040 |
0.0 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 2 |
Hb_034280_010 |
0.1447391734 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 3 |
Hb_028115_010 |
0.1456296096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000413_230 |
0.1765154504 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 5 |
Hb_010872_040 |
0.2064420425 |
- |
- |
hypothetical protein F775_31993 [Aegilops tauschii] |
| 6 |
Hb_173741_010 |
0.2079951216 |
- |
- |
PREDICTED: caffeoyl-CoA O-methyltransferase isoform X3 [Musa acuminata subsp. malaccensis] |
| 7 |
Hb_000679_200 |
0.210972578 |
- |
- |
EIN3-binding F box protein 1 [Theobroma cacao] |
| 8 |
Hb_032208_050 |
0.2171283698 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 9 |
Hb_018691_010 |
0.2182221027 |
- |
- |
- |
| 10 |
Hb_000130_060 |
0.2196779692 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 11 |
Hb_001900_060 |
0.2201581872 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 12 |
Hb_000818_080 |
0.223920633 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 13 |
Hb_000152_460 |
0.2246360088 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 14 |
Hb_060391_010 |
0.22553814 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_003050_100 |
0.2259927781 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |
| 16 |
Hb_004724_120 |
0.2292898568 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 17 |
Hb_000029_440 |
0.2306809577 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit isoform X2 [Vitis vinifera] |
| 18 |
Hb_001124_110 |
0.2307608411 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_029510_100 |
0.231908594 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 20 |
Hb_000000_110 |
0.2364699925 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |