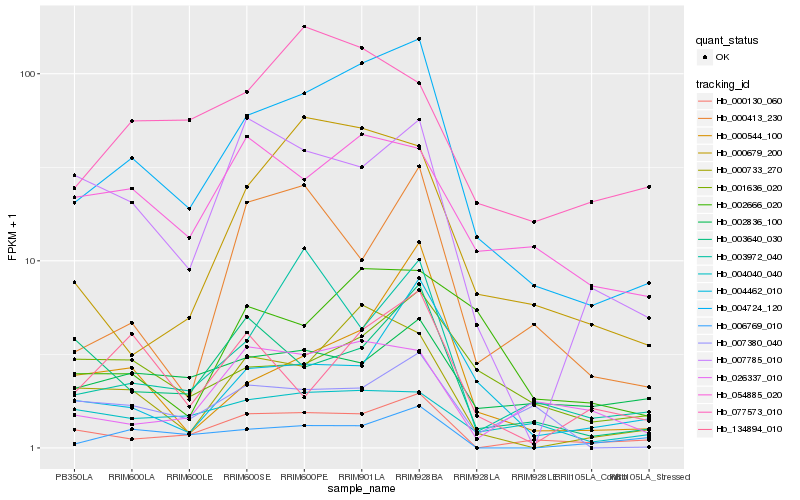
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004724_120 |
0.0 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 2 |
Hb_000733_270 |
0.177600916 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-alpha catalytic subunit [Dasypus novemcinctus] |
| 3 |
Hb_000130_060 |
0.1881678861 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 4 |
Hb_001636_020 |
0.2075674849 |
- |
- |
Serine/threonine-protein kinase PBS1 [Theobroma cacao] |
| 5 |
Hb_000679_200 |
0.2101140351 |
- |
- |
EIN3-binding F box protein 1 [Theobroma cacao] |
| 6 |
Hb_002666_020 |
0.2135865298 |
- |
- |
VAMP/SYNAPTOBREVIN-ASSOCIATED protein 27-1 [Populus trichocarpa] |
| 7 |
Hb_054885_020 |
0.2173639693 |
- |
- |
- |
| 8 |
Hb_004462_010 |
0.2176107958 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 9 |
Hb_000544_100 |
0.219229515 |
- |
- |
hypothetical protein JCGZ_03636 [Jatropha curcas] |
| 10 |
Hb_004040_040 |
0.2218194776 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 11 |
Hb_026337_010 |
0.2238371458 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 12 |
Hb_134894_010 |
0.2238892312 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 13 |
Hb_002836_100 |
0.2250116061 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007380_040 |
0.2258969585 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 15 |
Hb_077573_010 |
0.2270610213 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_007785_010 |
0.2274809552 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |
| 17 |
Hb_003640_030 |
0.2277104519 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003972_040 |
0.2292898568 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 19 |
Hb_000413_230 |
0.2293250106 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 20 |
Hb_006769_010 |
0.2297705253 |
- |
- |
hypothetical protein JCGZ_00291 [Jatropha curcas] |