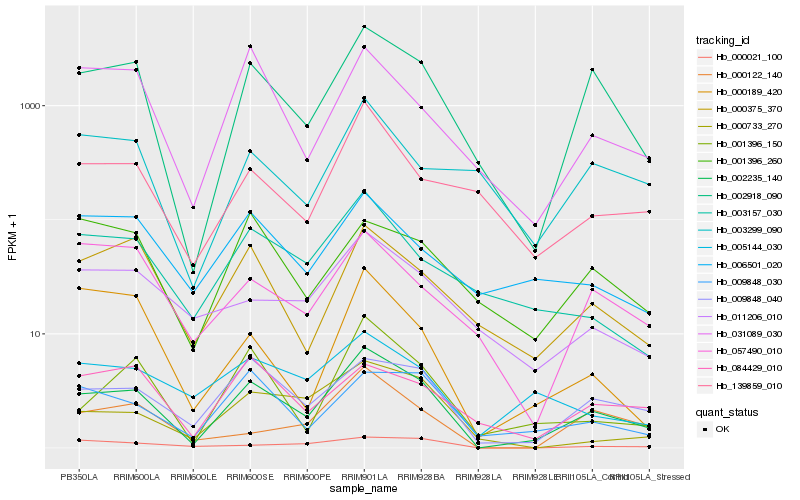
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002918_090 |
0.1499171565 |
- |
- |
PREDICTED: auxin-repressed 12.5 kDa protein-like [Nelumbo nucifera] |
| 3 |
Hb_009848_040 |
0.1688083656 |
- |
- |
hypothetical protein B456_009G349600 [Gossypium raimondii] |
| 4 |
Hb_001396_150 |
0.2005561064 |
transcription factor |
TF Family: M-type |
hypothetical protein POPTR_0001s13660g [Populus trichocarpa] |
| 5 |
Hb_009848_030 |
0.2037491421 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |
| 6 |
Hb_000375_370 |
0.209603638 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_031089_030 |
0.2158758476 |
- |
- |
ubiquitin-conjugating enzyme E2 28 [Arabidopsis thaliana] |
| 8 |
Hb_000733_270 |
0.2214105367 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-alpha catalytic subunit [Dasypus novemcinctus] |
| 9 |
Hb_003157_030 |
0.2217293474 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Nicotiana sylvestris] |
| 10 |
Hb_000122_140 |
0.2218968588 |
- |
- |
- |
| 11 |
Hb_084429_010 |
0.2228123407 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Populus euphratica] |
| 12 |
Hb_139859_010 |
0.2230587945 |
- |
- |
PREDICTED: cell division control protein 48 homolog D-like [Nicotiana sylvestris] |
| 13 |
Hb_005144_030 |
0.2268811903 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 14 |
Hb_011206_010 |
0.2300402912 |
- |
- |
PREDICTED: something about silencing protein 10 isoform X2 [Vitis vinifera] |
| 15 |
Hb_001396_260 |
0.2315144504 |
- |
- |
PREDICTED: F-box only protein 6 [Jatropha curcas] |
| 16 |
Hb_000189_420 |
0.2316609703 |
- |
- |
PREDICTED: uncharacterized protein LOC105647665 [Jatropha curcas] |
| 17 |
Hb_006501_020 |
0.232658378 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 18 |
Hb_000021_100 |
0.2334692339 |
- |
- |
- |
| 19 |
Hb_057490_010 |
0.2353723588 |
- |
- |
PREDICTED: mannose-1-phosphate guanyltransferase alpha [Jatropha curcas] |
| 20 |
Hb_003299_090 |
0.2394239491 |
- |
- |
hypothetical protein PRUPE_ppa011647mg [Prunus persica] |