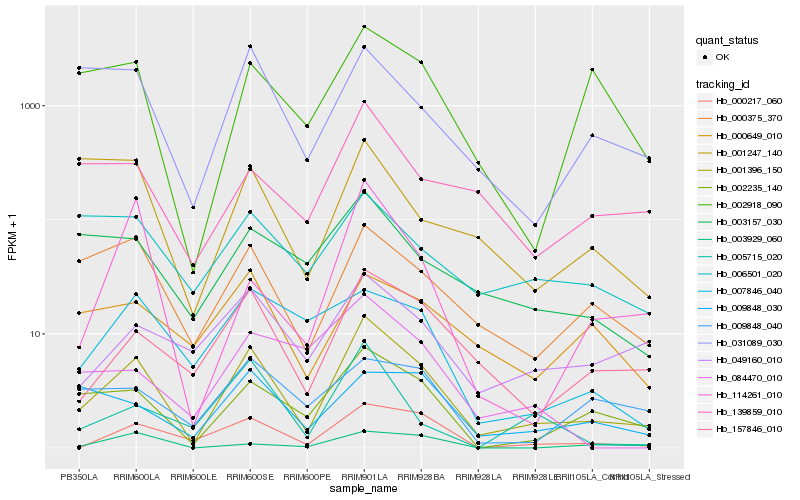
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001396_150 |
0.0 |
transcription factor |
TF Family: M-type |
hypothetical protein POPTR_0001s13660g [Populus trichocarpa] |
| 2 |
Hb_157846_010 |
0.1983827022 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 3 |
Hb_002235_140 |
0.2005561064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000217_060 |
0.220843519 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000375_370 |
0.2240150709 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_049160_010 |
0.2388090521 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 7 |
Hb_114261_010 |
0.252959285 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 8 |
Hb_031089_030 |
0.2530744967 |
- |
- |
ubiquitin-conjugating enzyme E2 28 [Arabidopsis thaliana] |
| 9 |
Hb_139859_010 |
0.2535410511 |
- |
- |
PREDICTED: cell division control protein 48 homolog D-like [Nicotiana sylvestris] |
| 10 |
Hb_005715_020 |
0.2561046046 |
- |
- |
PREDICTED: ras-related protein RABC2a-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_002918_090 |
0.2567606897 |
- |
- |
PREDICTED: auxin-repressed 12.5 kDa protein-like [Nelumbo nucifera] |
| 12 |
Hb_003157_030 |
0.2599902369 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Nicotiana sylvestris] |
| 13 |
Hb_009848_030 |
0.2670037841 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |
| 14 |
Hb_000649_010 |
0.2676262406 |
- |
- |
PREDICTED: adagio protein 3 [Jatropha curcas] |
| 15 |
Hb_001247_140 |
0.2736355109 |
- |
- |
- |
| 16 |
Hb_007846_040 |
0.2736786688 |
- |
- |
PREDICTED: uncharacterized protein LOC105646691 [Jatropha curcas] |
| 17 |
Hb_009848_040 |
0.275252703 |
- |
- |
hypothetical protein B456_009G349600 [Gossypium raimondii] |
| 18 |
Hb_003929_060 |
0.2756909728 |
- |
- |
PREDICTED: uncharacterized protein ECU03_1610 [Jatropha curcas] |
| 19 |
Hb_006501_020 |
0.2762613683 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 20 |
Hb_084470_010 |
0.2765101752 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |