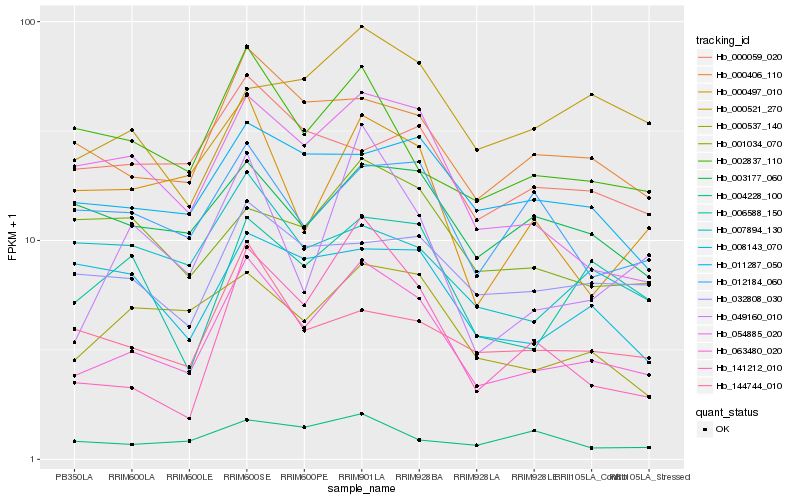
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_063480_020 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_054885_020 |
0.1317122401 |
- |
- |
- |
| 3 |
Hb_000406_110 |
0.1343477915 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 4 |
Hb_000537_140 |
0.1406089599 |
- |
- |
lupus la ribonucleoprotein, putative [Ricinus communis] |
| 5 |
Hb_006588_150 |
0.1442778047 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 6 |
Hb_002837_110 |
0.1455640972 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_049160_010 |
0.1516232346 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 8 |
Hb_000497_010 |
0.1553551488 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR2 [Jatropha curcas] |
| 9 |
Hb_011287_050 |
0.1556587555 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 10 |
Hb_032808_030 |
0.1560857724 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_141212_010 |
0.1565514513 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 12 |
Hb_003177_060 |
0.158569588 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_144744_010 |
0.1589810372 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 14 |
Hb_012184_060 |
0.160201227 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007894_130 |
0.1664920055 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 16 |
Hb_001034_070 |
0.1667618442 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_008143_070 |
0.1695235141 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 18 |
Hb_000521_270 |
0.1698614336 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 19 |
Hb_004228_100 |
0.1700966143 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 20 |
Hb_000059_020 |
0.170338131 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |