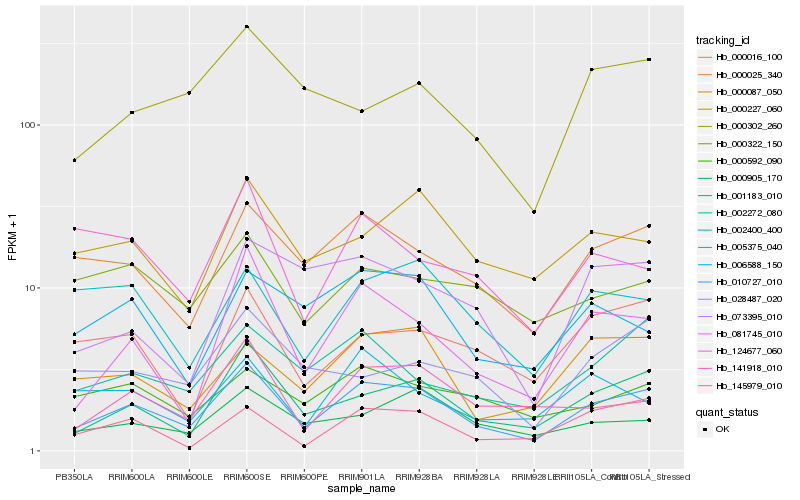
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010727_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 2 |
Hb_001183_010 |
0.1279627033 |
- |
- |
- |
| 3 |
Hb_000025_340 |
0.1412545844 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350 [Jatropha curcas] |
| 4 |
Hb_081745_010 |
0.1459531689 |
- |
- |
hypothetical protein CISIN_1g030953mg [Citrus sinensis] |
| 5 |
Hb_141918_010 |
0.159729207 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 6 |
Hb_000087_050 |
0.1628670941 |
- |
- |
hypothetical protein JCGZ_16227 [Jatropha curcas] |
| 7 |
Hb_002272_080 |
0.1667769142 |
- |
- |
- |
| 8 |
Hb_000227_060 |
0.1683375997 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 9 |
Hb_000905_170 |
0.1685788506 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 10 |
Hb_002400_400 |
0.1726087024 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 11 |
Hb_000016_100 |
0.1732038152 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP19 [Jatropha curcas] |
| 12 |
Hb_000592_090 |
0.1740413035 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18020 [Jatropha curcas] |
| 13 |
Hb_145979_010 |
0.1745728031 |
- |
- |
PREDICTED: uncharacterized protein LOC105172316 [Sesamum indicum] |
| 14 |
Hb_006588_150 |
0.1748981834 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 15 |
Hb_005375_040 |
0.1863146379 |
- |
- |
dicer-1, putative [Ricinus communis] |
| 16 |
Hb_073395_010 |
0.1870138333 |
- |
- |
- |
| 17 |
Hb_000302_260 |
0.1872004118 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 18 |
Hb_124677_060 |
0.1877496832 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000322_150 |
0.1885457652 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_028487_020 |
0.189207176 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |