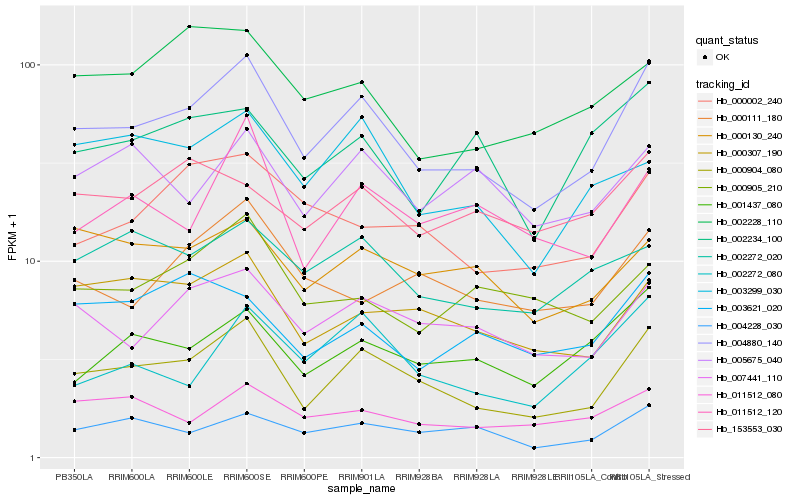
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004880_140 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 2 |
Hb_000904_080 |
0.0651369056 |
- |
- |
alpha-amylase-like protein [Vitis labrusca x Vitis vinifera] |
| 3 |
Hb_007441_110 |
0.11301876 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At3g58900-like [Jatropha curcas] |
| 4 |
Hb_004228_030 |
0.1232088196 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_001437_080 |
0.1296948674 |
transcription factor |
TF Family: TCP |
hypothetical protein JCGZ_05472 [Jatropha curcas] |
| 6 |
Hb_002272_020 |
0.1329428296 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 7 |
Hb_000307_190 |
0.1333513744 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g20300, mitochondrial-like [Populus euphratica] |
| 8 |
Hb_011512_080 |
0.1342329526 |
- |
- |
hypothetical protein JCGZ_06424 [Jatropha curcas] |
| 9 |
Hb_000002_240 |
0.1352174081 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 10 |
Hb_000130_240 |
0.1358290965 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 11 |
Hb_002234_100 |
0.1363336608 |
- |
- |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 12 |
Hb_002272_080 |
0.1373221184 |
- |
- |
- |
| 13 |
Hb_002228_110 |
0.1409850799 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 14 |
Hb_000111_180 |
0.1412021959 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 15 |
Hb_003299_030 |
0.1417864576 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 16 |
Hb_011512_120 |
0.1418407822 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 17 |
Hb_005675_040 |
0.1423718913 |
transcription factor |
TF Family: ERF |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000905_210 |
0.1439875677 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 19 |
Hb_153553_030 |
0.1441606156 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 20 |
Hb_003621_020 |
0.1469262226 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |