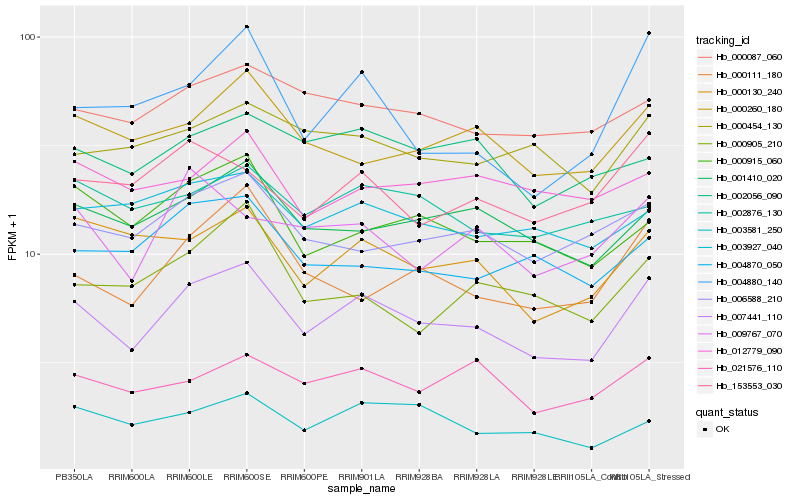
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_110 |
0.0 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At3g58900-like [Jatropha curcas] |
| 2 |
Hb_001410_020 |
0.102669006 |
- |
- |
cdk8, putative [Ricinus communis] |
| 3 |
Hb_000260_180 |
0.1067821242 |
- |
- |
PREDICTED: KH domain-containing protein At2g38610 [Jatropha curcas] |
| 4 |
Hb_006588_210 |
0.1067856364 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 5 |
Hb_000130_240 |
0.1067923336 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 6 |
Hb_021576_110 |
0.1073404632 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 [Jatropha curcas] |
| 7 |
Hb_000087_060 |
0.1100031423 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 8 |
Hb_000915_060 |
0.1105723949 |
- |
- |
PREDICTED: uncharacterized protein LOC105628864 [Jatropha curcas] |
| 9 |
Hb_000111_180 |
0.1106001864 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 10 |
Hb_003927_040 |
0.1123203837 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 11 |
Hb_004880_140 |
0.11301876 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 12 |
Hb_003581_250 |
0.1151164721 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 13 |
Hb_002876_130 |
0.1158271482 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 14 |
Hb_153553_030 |
0.1160006614 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 15 |
Hb_004870_050 |
0.1163291862 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 16 |
Hb_002056_090 |
0.1165892084 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 17 |
Hb_000454_130 |
0.1167129939 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_009767_070 |
0.119211172 |
- |
- |
PREDICTED: uncharacterized protein LOC105631980 [Jatropha curcas] |
| 19 |
Hb_000905_210 |
0.1221634216 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 20 |
Hb_012779_090 |
0.1223305207 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |