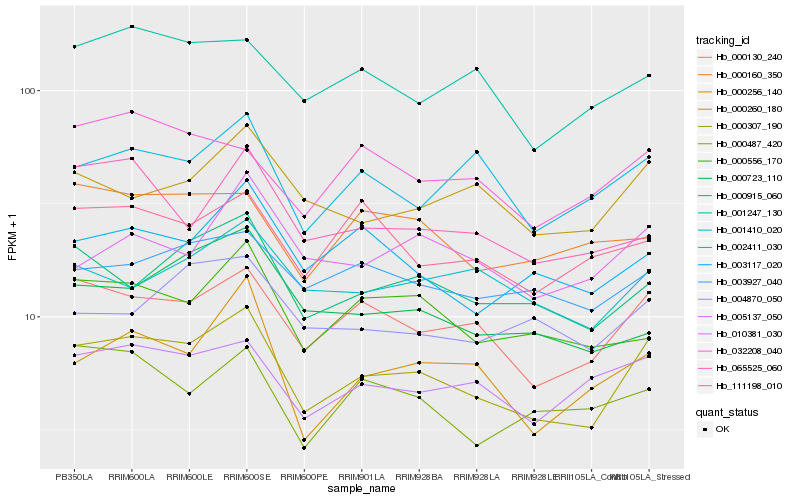
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000307_190 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g20300, mitochondrial-like [Populus euphratica] |
| 2 |
Hb_000130_240 |
0.0774858628 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 3 |
Hb_000915_060 |
0.0989424685 |
- |
- |
PREDICTED: uncharacterized protein LOC105628864 [Jatropha curcas] |
| 4 |
Hb_005137_050 |
0.1006957577 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 5 |
Hb_000256_140 |
0.1075241854 |
- |
- |
PREDICTED: uncharacterized protein LOC105637585 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002411_030 |
0.1092888743 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor homolog [Jatropha curcas] |
| 7 |
Hb_003117_020 |
0.1101981087 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE6 [Jatropha curcas] |
| 8 |
Hb_000260_180 |
0.1108495575 |
- |
- |
PREDICTED: KH domain-containing protein At2g38610 [Jatropha curcas] |
| 9 |
Hb_000556_170 |
0.1122856623 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001247_130 |
0.112471695 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 11 |
Hb_003927_040 |
0.112694013 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 12 |
Hb_001410_020 |
0.1157003095 |
- |
- |
cdk8, putative [Ricinus communis] |
| 13 |
Hb_111198_010 |
0.1161971725 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_010381_030 |
0.1166270809 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 15 |
Hb_032208_040 |
0.1169967273 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL33 [Populus euphratica] |
| 16 |
Hb_065525_060 |
0.1185425308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004870_050 |
0.1189105127 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 18 |
Hb_000160_350 |
0.1194545292 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 19 |
Hb_000723_110 |
0.1197527458 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 20 |
Hb_000487_420 |
0.1199345999 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g04390 [Jatropha curcas] |