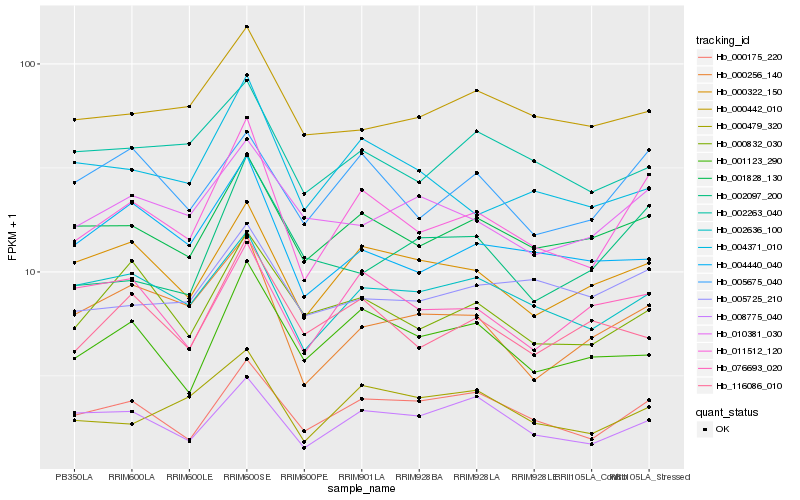
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011512_120 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 2 |
Hb_001828_130 |
0.1049642725 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 3 |
Hb_000175_220 |
0.1055872858 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 4 |
Hb_000322_150 |
0.1161617611 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002636_100 |
0.1197381452 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 6 |
Hb_005675_040 |
0.1201859234 |
transcription factor |
TF Family: ERF |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000256_140 |
0.1238462374 |
- |
- |
PREDICTED: uncharacterized protein LOC105637585 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000832_030 |
0.1240601746 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634915 [Jatropha curcas] |
| 9 |
Hb_000479_320 |
0.1270990376 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06140, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_004371_010 |
0.1273056439 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 11 |
Hb_001123_290 |
0.1282628435 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 12 |
Hb_008775_040 |
0.1286151987 |
- |
- |
Pentatricopeptide repeat (PPR) superfamily protein, putative [Theobroma cacao] |
| 13 |
Hb_010381_030 |
0.1304002077 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 14 |
Hb_004440_040 |
0.1307721379 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_005725_210 |
0.132580623 |
- |
- |
PREDICTED: RNA-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_076693_020 |
0.132754751 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 17 |
Hb_116086_010 |
0.1330099669 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 18 |
Hb_002263_040 |
0.1344530648 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 19 |
Hb_000442_010 |
0.1361316943 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 20 |
Hb_002097_200 |
0.1361345993 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |