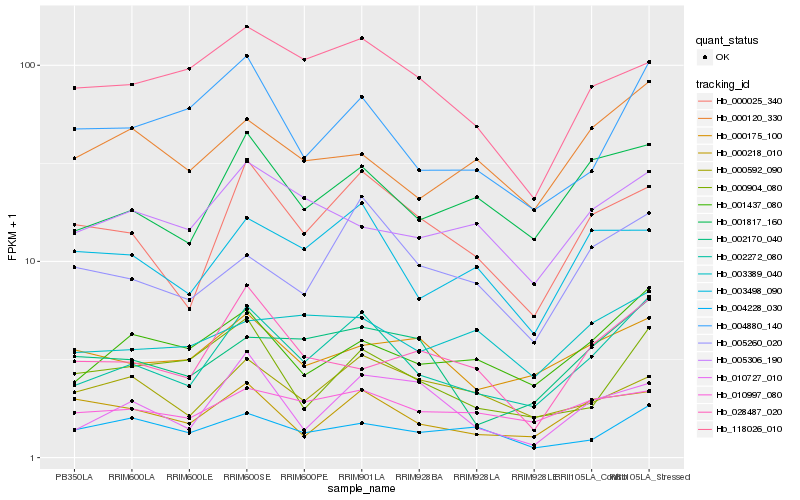
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002272_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_000025_340 |
0.1200422854 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350 [Jatropha curcas] |
| 3 |
Hb_001817_160 |
0.1239024282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001437_080 |
0.135748723 |
transcription factor |
TF Family: TCP |
hypothetical protein JCGZ_05472 [Jatropha curcas] |
| 5 |
Hb_004880_140 |
0.1373221184 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 6 |
Hb_005260_020 |
0.1453330233 |
- |
- |
PREDICTED: uncharacterized protein LOC105633789 [Jatropha curcas] |
| 7 |
Hb_010997_080 |
0.1478968521 |
transcription factor |
TF Family: mTERF |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_002170_040 |
0.1487424697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003498_090 |
0.1488387833 |
- |
- |
hypothetical protein RCOM_0867850 [Ricinus communis] |
| 10 |
Hb_000218_010 |
0.1583427341 |
- |
- |
- |
| 11 |
Hb_000904_080 |
0.1604593326 |
- |
- |
alpha-amylase-like protein [Vitis labrusca x Vitis vinifera] |
| 12 |
Hb_000175_100 |
0.1617236542 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 13 |
Hb_005306_190 |
0.1622897764 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 14 |
Hb_118026_010 |
0.162830761 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 15 |
Hb_028487_020 |
0.1641971839 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 16 |
Hb_000120_330 |
0.16428626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004228_030 |
0.1645921199 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_003389_040 |
0.1652246568 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |
| 19 |
Hb_000592_090 |
0.1654707195 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18020 [Jatropha curcas] |
| 20 |
Hb_010727_010 |
0.1667769142 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |