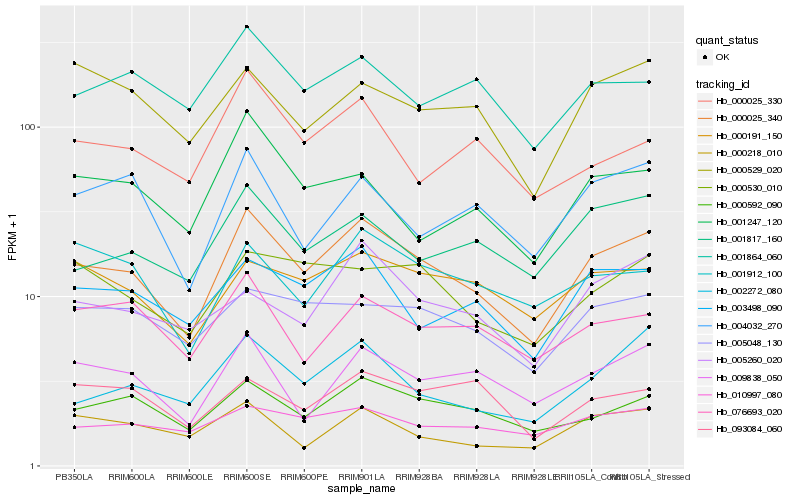
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_340 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350 [Jatropha curcas] |
| 2 |
Hb_000592_090 |
0.1085368541 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18020 [Jatropha curcas] |
| 3 |
Hb_003498_090 |
0.1163471049 |
- |
- |
hypothetical protein RCOM_0867850 [Ricinus communis] |
| 4 |
Hb_002272_080 |
0.1200422854 |
- |
- |
- |
| 5 |
Hb_009838_050 |
0.1241984684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000530_010 |
0.1249084524 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03 [Jatropha curcas] |
| 7 |
Hb_001817_160 |
0.1251163202 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000191_150 |
0.1259451952 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 9 |
Hb_005048_130 |
0.1265793346 |
- |
- |
hypothetical protein POPTR_0004s03700g [Populus trichocarpa] |
| 10 |
Hb_010997_080 |
0.1290934413 |
transcription factor |
TF Family: mTERF |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_076693_020 |
0.1300061865 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 12 |
Hb_001864_060 |
0.1317264649 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 13 |
Hb_004032_270 |
0.1324261017 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_005260_020 |
0.1333467512 |
- |
- |
PREDICTED: uncharacterized protein LOC105633789 [Jatropha curcas] |
| 15 |
Hb_001912_100 |
0.133944541 |
- |
- |
PREDICTED: uncharacterized protein LOC105636609 [Jatropha curcas] |
| 16 |
Hb_000218_010 |
0.1364734339 |
- |
- |
- |
| 17 |
Hb_000529_020 |
0.1368184708 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 18 |
Hb_001247_120 |
0.1377552423 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 19 |
Hb_000025_330 |
0.1385618762 |
- |
- |
hypothetical protein JCGZ_22759 [Jatropha curcas] |
| 20 |
Hb_093084_060 |
0.1389417817 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At1g64310 [Jatropha curcas] |