| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
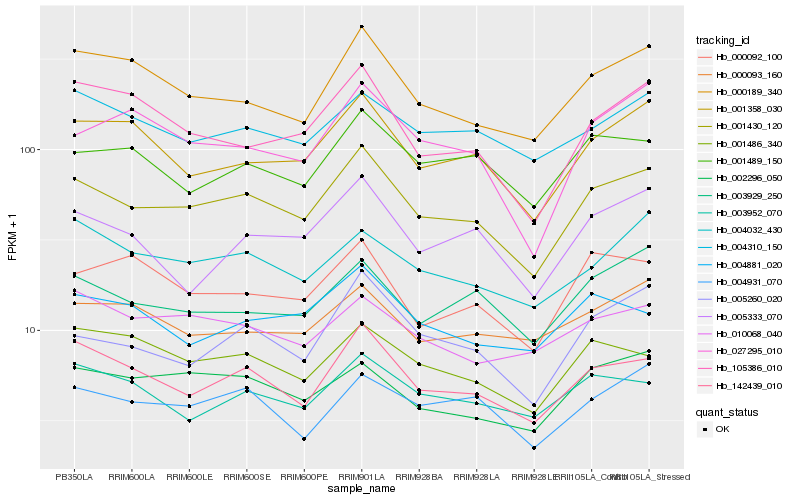
Hb_001430_120 |
0.0 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 2 |
Hb_142439_010 |
0.0652766385 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 3 |
Hb_001358_030 |
0.080650261 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 4 |
Hb_000189_340 |
0.0814812214 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |
| 5 |
Hb_005260_020 |
0.0831309879 |
- |
- |
PREDICTED: uncharacterized protein LOC105633789 [Jatropha curcas] |
| 6 |
Hb_002296_050 |
0.0857207598 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 7 |
Hb_004310_150 |
0.0868513398 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 8 |
Hb_003929_250 |
0.0873264048 |
- |
- |
PREDICTED: proteasome assembly chaperone 4 [Jatropha curcas] |
| 9 |
Hb_004931_070 |
0.0874351024 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001489_150 |
0.0877290989 |
- |
- |
PREDICTED: uncharacterized protein LOC105123623 isoform X2 [Populus euphratica] |
| 11 |
Hb_004032_430 |
0.0888727425 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |
| 12 |
Hb_005333_070 |
0.0906359694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001486_340 |
0.0912024235 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 14 |
Hb_003952_070 |
0.0930235519 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_105386_010 |
0.0943635003 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 16 |
Hb_027295_010 |
0.0944625479 |
- |
- |
40S ribosomal protein S7, putative [Ricinus communis] |
| 17 |
Hb_000092_100 |
0.094629623 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 18 |
Hb_000093_160 |
0.0955839882 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_010068_040 |
0.0958366227 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 20 |
Hb_004881_020 |
0.0960036535 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |