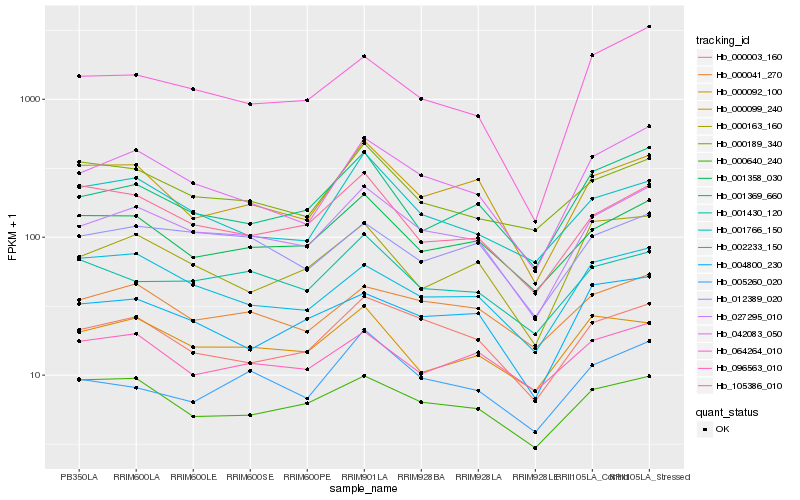
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027295_010 |
0.0 |
- |
- |
40S ribosomal protein S7, putative [Ricinus communis] |
| 2 |
Hb_042083_050 |
0.0680436454 |
- |
- |
60S ribosomal protein L11, putative [Ricinus communis] |
| 3 |
Hb_000003_160 |
0.0729986774 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 4 |
Hb_001358_030 |
0.0774310847 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 5 |
Hb_001369_660 |
0.0884508915 |
- |
- |
hypothetical protein B456_001G174200, partial [Gossypium raimondii] |
| 6 |
Hb_012389_020 |
0.0896473199 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |
| 7 |
Hb_000099_240 |
0.0897943484 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 8 |
Hb_000041_270 |
0.0905602612 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 9 |
Hb_000640_240 |
0.0920847138 |
- |
- |
carboxypeptidase regulatory region-containingprotein, putative [Ricinus communis] |
| 10 |
Hb_001430_120 |
0.0944625479 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 11 |
Hb_105386_010 |
0.0960526066 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 12 |
Hb_004800_230 |
0.0970431241 |
- |
- |
PREDICTED: MIP18 family protein At1g68310 [Jatropha curcas] |
| 13 |
Hb_005260_020 |
0.0975597806 |
- |
- |
PREDICTED: uncharacterized protein LOC105633789 [Jatropha curcas] |
| 14 |
Hb_001766_150 |
0.0988880543 |
- |
- |
PREDICTED: 40S ribosomal protein S6 [Jatropha curcas] |
| 15 |
Hb_002233_150 |
0.0990830856 |
- |
- |
PREDICTED: uncharacterized protein LOC105633093 [Jatropha curcas] |
| 16 |
Hb_000189_340 |
0.0991057693 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |
| 17 |
Hb_000163_160 |
0.0992024932 |
- |
- |
Putative small nuclear ribonucleoprotein Sm D2 [Glycine soja] |
| 18 |
Hb_064264_010 |
0.0999999276 |
- |
- |
hypothetical protein [Zea mays] |
| 19 |
Hb_000092_100 |
0.1020937936 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 20 |
Hb_096563_010 |
0.1022157237 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Prunus mume] |