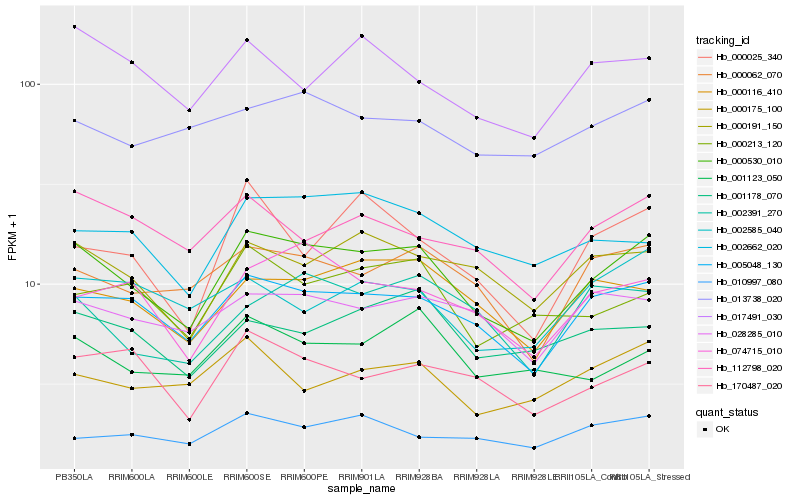
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000530_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03 [Jatropha curcas] |
| 2 |
Hb_005048_130 |
0.0809349207 |
- |
- |
hypothetical protein POPTR_0004s03700g [Populus trichocarpa] |
| 3 |
Hb_000062_070 |
0.1028779311 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 4 |
Hb_000191_150 |
0.1055415511 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 5 |
Hb_000116_410 |
0.1090766241 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 6 |
Hb_074715_010 |
0.1105127342 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_112798_020 |
0.1121792268 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 8 |
Hb_170487_020 |
0.1162953237 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 9 |
Hb_028285_010 |
0.1172750163 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 10 |
Hb_002662_020 |
0.1180795051 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 11 |
Hb_001123_050 |
0.1181334225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000213_120 |
0.118516532 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_001178_070 |
0.1206922683 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein FBL11 [Jatropha curcas] |
| 14 |
Hb_017491_030 |
0.1209456552 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 15 |
Hb_013738_020 |
0.1211225231 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 16 |
Hb_002391_270 |
0.1212914627 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 17 |
Hb_000175_100 |
0.1236357584 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000025_340 |
0.1249084524 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350 [Jatropha curcas] |
| 19 |
Hb_002585_040 |
0.1257523249 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 20 |
Hb_010997_080 |
0.1262643778 |
transcription factor |
TF Family: mTERF |
unnamed protein product [Vitis vinifera] |