| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
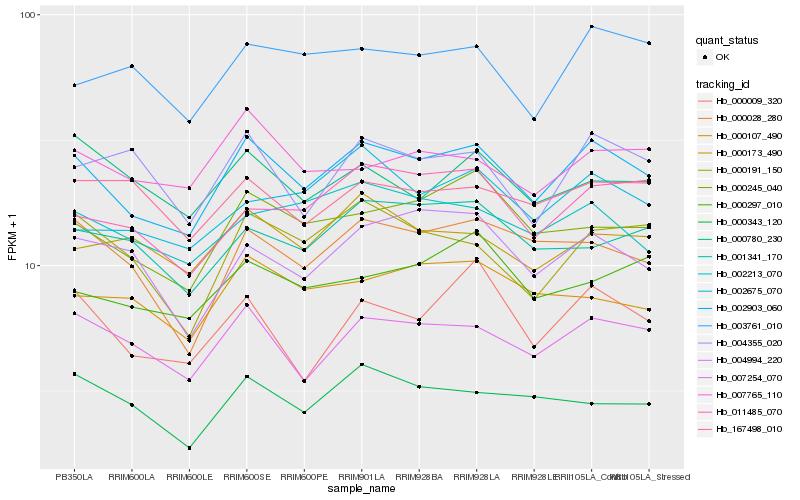
Hb_002903_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649505 [Jatropha curcas] |
| 2 |
Hb_007254_070 |
0.0652461333 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000245_040 |
0.0766377259 |
- |
- |
PREDICTED: uncharacterized protein LOC105631550 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000028_280 |
0.0804548317 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 5 |
Hb_000191_150 |
0.0821559974 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 6 |
Hb_002213_070 |
0.0822836672 |
- |
- |
epsilon-adaptin family protein [Populus trichocarpa] |
| 7 |
Hb_011485_070 |
0.0837239046 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 8 |
Hb_000173_490 |
0.0839865147 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000343_120 |
0.0853066103 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16010 [Jatropha curcas] |
| 10 |
Hb_004994_220 |
0.0853155659 |
- |
- |
PREDICTED: K(+) efflux antiporter 2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_007765_110 |
0.0855801838 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 12 |
Hb_001341_170 |
0.0875650973 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 13 |
Hb_003761_010 |
0.0875901995 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_004355_020 |
0.0878818769 |
- |
- |
PREDICTED: lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000009_320 |
0.0879422886 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66631 [Jatropha curcas] |
| 16 |
Hb_000107_490 |
0.0880039191 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 17 |
Hb_002675_070 |
0.0903171888 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 18 |
Hb_167498_010 |
0.0904346364 |
- |
- |
PREDICTED: WPP domain-interacting protein 1-like [Jatropha curcas] |
| 19 |
Hb_000780_230 |
0.0910747331 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 20 |
Hb_000297_010 |
0.0910963979 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |