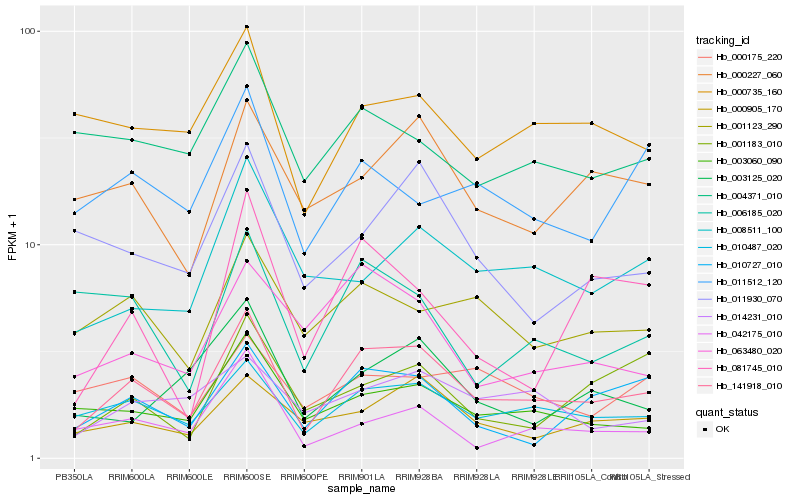
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_141918_010 |
0.0 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 2 |
Hb_010487_020 |
0.1190565858 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 3 |
Hb_010727_010 |
0.159729207 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 4 |
Hb_003060_090 |
0.1609480773 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 5 |
Hb_000905_170 |
0.1617814333 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 6 |
Hb_001123_290 |
0.1706121786 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 7 |
Hb_000227_060 |
0.1716006755 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 8 |
Hb_063480_020 |
0.1772356965 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000735_160 |
0.1784707356 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |
| 10 |
Hb_014231_010 |
0.1795169863 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 11 |
Hb_008511_100 |
0.1803226827 |
- |
- |
hypothetical protein POPTR_0009s11860g [Populus trichocarpa] |
| 12 |
Hb_042175_010 |
0.1807825337 |
- |
- |
hypothetical protein CISIN_1g0016721mg, partial [Citrus sinensis] |
| 13 |
Hb_011930_070 |
0.1843656543 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 14 |
Hb_006185_020 |
0.1878977673 |
- |
- |
PREDICTED: midasin [Jatropha curcas] |
| 15 |
Hb_004371_010 |
0.1881043875 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_011512_120 |
0.188536773 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 17 |
Hb_081745_010 |
0.1899299011 |
- |
- |
hypothetical protein CISIN_1g030953mg [Citrus sinensis] |
| 18 |
Hb_001183_010 |
0.19025962 |
- |
- |
- |
| 19 |
Hb_003125_020 |
0.1910853069 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_000175_220 |
0.1916671429 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |