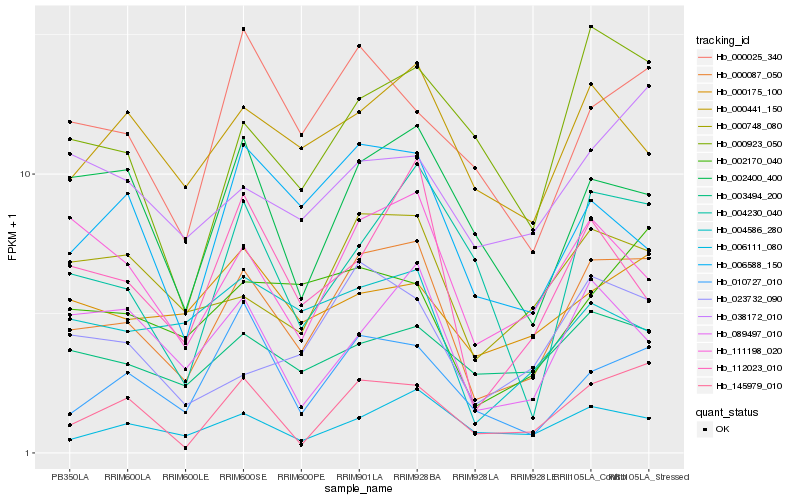
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000087_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_16227 [Jatropha curcas] |
| 2 |
Hb_000748_080 |
0.1447527424 |
- |
- |
hypothetical protein OsI_28173 [Oryza sativa Indica Group] |
| 3 |
Hb_006111_080 |
0.1479780243 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 4 |
Hb_145979_010 |
0.1504301988 |
- |
- |
PREDICTED: uncharacterized protein LOC105172316 [Sesamum indicum] |
| 5 |
Hb_111198_020 |
0.1546097527 |
- |
- |
hypothetical protein JCGZ_06907 [Jatropha curcas] |
| 6 |
Hb_023732_090 |
0.1556140205 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_003494_200 |
0.1576469444 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15-like [Jatropha curcas] |
| 8 |
Hb_002170_040 |
0.1588286887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000923_050 |
0.1606151917 |
- |
- |
PREDICTED: transcription factor bHLH113 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002400_400 |
0.1608739597 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 11 |
Hb_000441_150 |
0.1615729267 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_000025_340 |
0.1623192524 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350 [Jatropha curcas] |
| 13 |
Hb_010727_010 |
0.1628670941 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 14 |
Hb_006588_150 |
0.1681398443 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 15 |
Hb_089497_010 |
0.1698322916 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_112023_010 |
0.1702205715 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 17 |
Hb_004230_040 |
0.1710226893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_038172_010 |
0.1731295335 |
desease resistance |
Gene Name: AAA |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 19 |
Hb_000175_100 |
0.1736263535 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 20 |
Hb_004586_280 |
0.1737021125 |
- |
- |
PREDICTED: GEM-like protein 1 [Jatropha curcas] |