| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
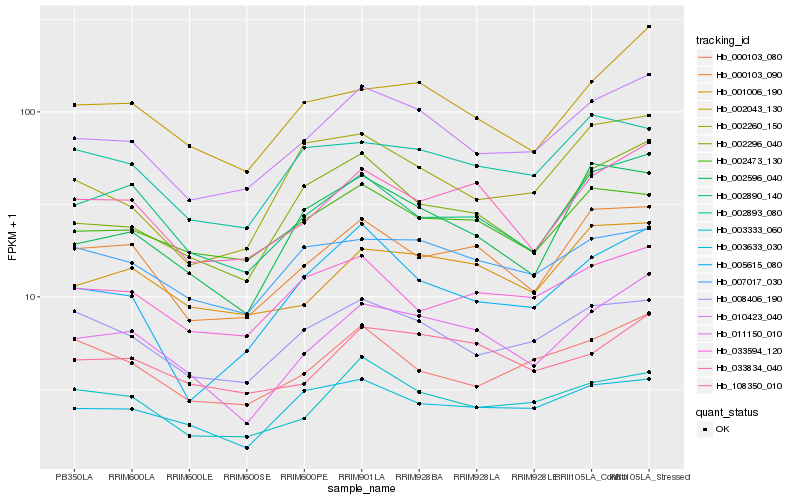
Hb_010423_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_17463 [Jatropha curcas] |
| 2 |
Hb_008406_190 |
0.0763610669 |
- |
- |
PREDICTED: RNA polymerase II subunit 5-mediating protein homolog isoform X2 [Jatropha curcas] |
| 3 |
Hb_003333_060 |
0.0791114721 |
- |
- |
PREDICTED: uncharacterized protein LOC105628461 [Jatropha curcas] |
| 4 |
Hb_002296_040 |
0.0840481328 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 4 [Jatropha curcas] |
| 5 |
Hb_005615_080 |
0.0859082301 |
- |
- |
PREDICTED: uncharacterized protein LOC105649609 [Jatropha curcas] |
| 6 |
Hb_000103_090 |
0.0876325306 |
- |
- |
PREDICTED: protein RDM1 [Jatropha curcas] |
| 7 |
Hb_000103_080 |
0.0892092374 |
- |
- |
PREDICTED: ATP-dependent RNA helicase SUV3, mitochondrial [Jatropha curcas] |
| 8 |
Hb_011150_010 |
0.0908050324 |
- |
- |
PREDICTED: uncharacterized RNA-binding protein C1827.05c [Jatropha curcas] |
| 9 |
Hb_001006_190 |
0.0910766638 |
- |
- |
PREDICTED: uncharacterized protein LOC105634201 [Jatropha curcas] |
| 10 |
Hb_033834_040 |
0.0918739345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002890_140 |
0.0927036706 |
- |
- |
PREDICTED: uncharacterized protein LOC100263805 [Vitis vinifera] |
| 12 |
Hb_002260_150 |
0.0928934251 |
- |
- |
PREDICTED: uncharacterized protein LOC105647187 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002473_130 |
0.0932688548 |
- |
- |
PREDICTED: THO complex subunit 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002043_130 |
0.0934218929 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 15 |
Hb_003633_030 |
0.094863249 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 16 |
Hb_108350_010 |
0.0959456195 |
- |
- |
GTP-binding protein erg, putative [Ricinus communis] |
| 17 |
Hb_033594_120 |
0.0959586585 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 18 |
Hb_007017_030 |
0.0962171161 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002596_040 |
0.0975308785 |
- |
- |
PREDICTED: uncharacterized protein LOC105645264 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002893_080 |
0.0981774052 |
- |
- |
PREDICTED: V-type proton ATPase subunit c''1 [Jatropha curcas] |