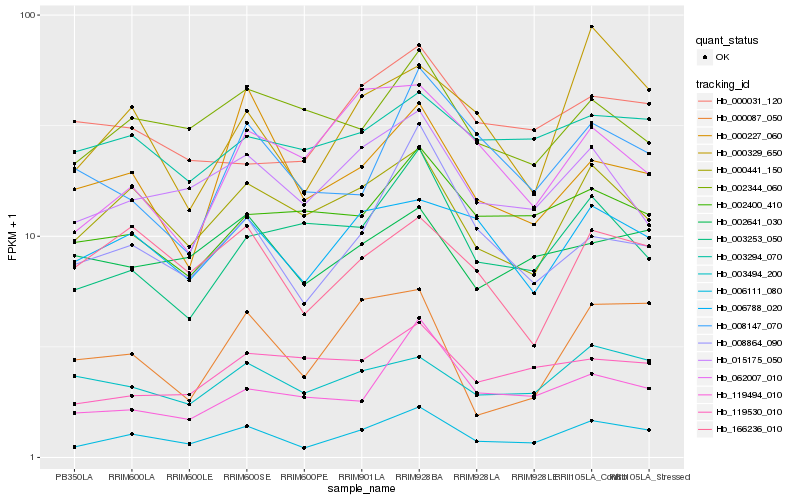
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006111_080 |
0.0 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 2 |
Hb_000441_150 |
0.1228859423 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_000329_650 |
0.1319817232 |
- |
- |
hypothetical protein POPTR_0014s056001g, partial [Populus trichocarpa] |
| 4 |
Hb_015175_050 |
0.1342734847 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10 isoform X3 [Jatropha curcas] |
| 5 |
Hb_008864_090 |
0.1349752943 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 6 |
Hb_006788_020 |
0.1390937639 |
- |
- |
hypothetical protein JCGZ_21864 [Jatropha curcas] |
| 7 |
Hb_119494_010 |
0.1461263962 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 8 |
Hb_166236_010 |
0.1474459033 |
- |
- |
- |
| 9 |
Hb_000087_050 |
0.1479780243 |
- |
- |
hypothetical protein JCGZ_16227 [Jatropha curcas] |
| 10 |
Hb_003253_050 |
0.1498250816 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 3 [Jatropha curcas] |
| 11 |
Hb_003494_200 |
0.1512664513 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15-like [Jatropha curcas] |
| 12 |
Hb_002344_060 |
0.1544159807 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 13 |
Hb_002400_410 |
0.1559259334 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |
| 14 |
Hb_000227_060 |
0.1566119703 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 15 |
Hb_008147_070 |
0.1576692039 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 16 |
Hb_002641_030 |
0.1589052222 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 17 |
Hb_000031_120 |
0.1606409285 |
- |
- |
PREDICTED: uncharacterized protein LOC105637488 [Jatropha curcas] |
| 18 |
Hb_062007_010 |
0.1619210555 |
- |
- |
PREDICTED: argininosuccinate lyase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003294_070 |
0.1620785784 |
- |
- |
hypothetical protein RCOM_1714550 [Ricinus communis] |
| 20 |
Hb_119530_010 |
0.1620964149 |
- |
- |
- |