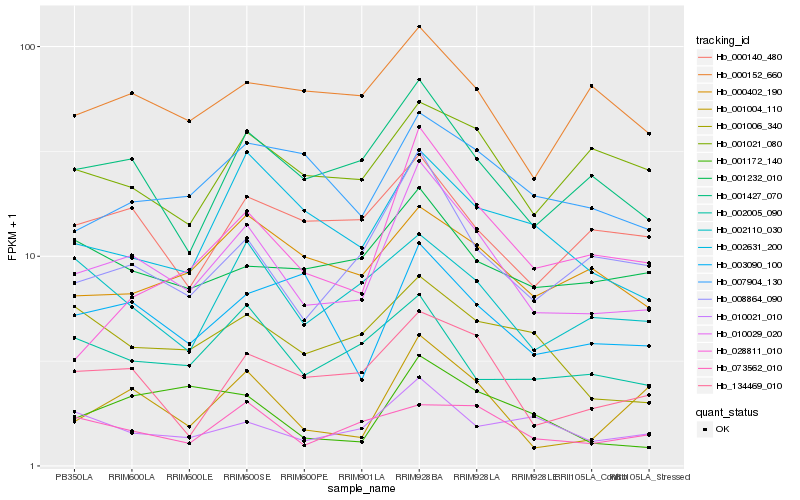
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010029_020 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 2 |
Hb_008864_090 |
0.1229629274 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 3 |
Hb_001427_070 |
0.1261636386 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 4 |
Hb_134469_010 |
0.1441692727 |
- |
- |
PREDICTED: protein argonaute 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001004_110 |
0.1492956534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007904_130 |
0.1575178898 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 7 |
Hb_010021_010 |
0.1580470185 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_002631_200 |
0.1593240937 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001172_140 |
0.1596326714 |
- |
- |
hypothetical protein POPTR_0014s17790g [Populus trichocarpa] |
| 10 |
Hb_000140_480 |
0.1599859095 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 11 |
Hb_001232_010 |
0.1600278264 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 12 |
Hb_002110_030 |
0.1613260621 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 13 |
Hb_000152_660 |
0.1618874862 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 14 |
Hb_000402_190 |
0.1618978021 |
- |
- |
PREDICTED: F-box protein At1g70590 [Jatropha curcas] |
| 15 |
Hb_001006_340 |
0.1625758374 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 16 |
Hb_003090_100 |
0.1632385639 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 17 |
Hb_028811_010 |
0.1640376524 |
- |
- |
PREDICTED: uncharacterized protein LOC105650178 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002005_090 |
0.1644924544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_073562_010 |
0.1652032705 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 20 |
Hb_001021_080 |
0.1652216974 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |