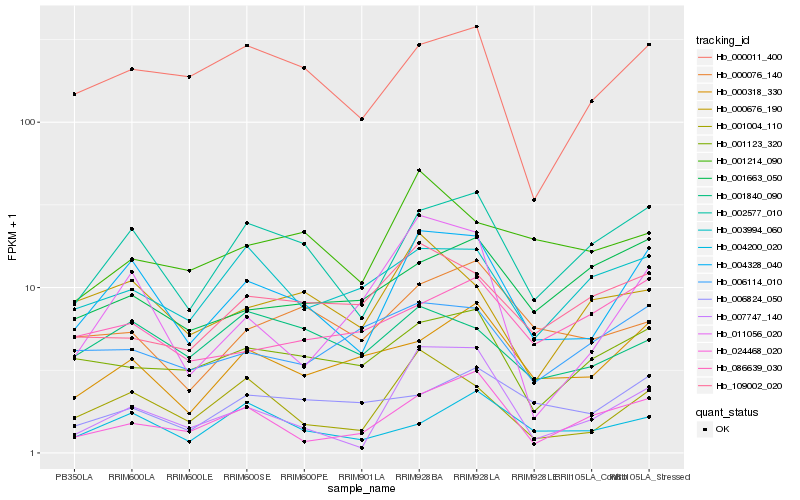
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004328_040 |
0.0 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 8 [Vitis vinifera] |
| 2 |
Hb_002577_010 |
0.1178036344 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 3 |
Hb_001004_110 |
0.1450116144 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000011_400 |
0.1678262201 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 4 [Hevea brasiliensis] |
| 5 |
Hb_000318_330 |
0.1695991571 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15630, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001123_320 |
0.1741707928 |
- |
- |
PREDICTED: uncharacterized protein LOC105647314 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000676_190 |
0.1822179972 |
- |
- |
hypothetical protein JCGZ_08279 [Jatropha curcas] |
| 8 |
Hb_007747_140 |
0.1830618981 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_004200_020 |
0.1862061276 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 10 |
Hb_001840_090 |
0.1866369338 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 11 |
Hb_003994_060 |
0.187293161 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 12 |
Hb_011056_020 |
0.1880000687 |
transcription factor |
TF Family: ERF |
AP2 domain transcription factor RAP2.3, putative [Ricinus communis] |
| 13 |
Hb_000076_140 |
0.1900520819 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_024468_020 |
0.1915140584 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 15 |
Hb_006114_010 |
0.1941821876 |
- |
- |
PREDICTED: uncharacterized protein LOC105629068 [Jatropha curcas] |
| 16 |
Hb_006824_050 |
0.1947496934 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_109002_020 |
0.1959818624 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 18 |
Hb_086639_030 |
0.196632315 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 19 |
Hb_001214_090 |
0.197049151 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 20 |
Hb_001663_050 |
0.19711747 |
- |
- |
PREDICTED: GPN-loop GTPase 3 [Jatropha curcas] |