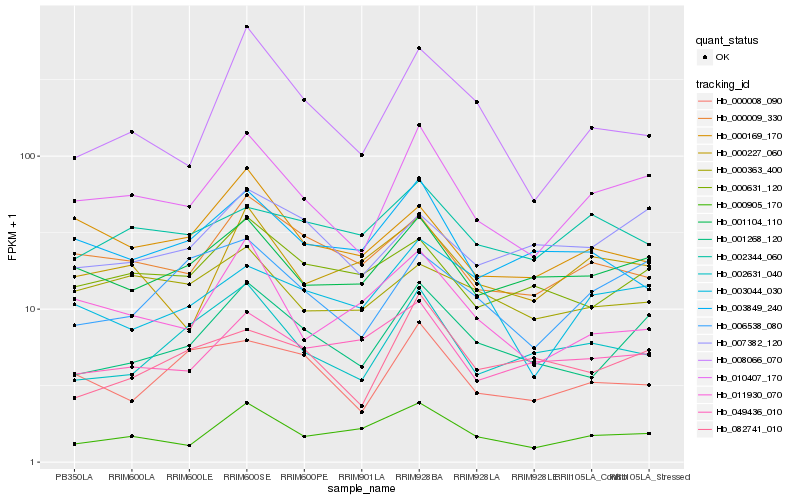
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010407_170 |
0.0 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 2 |
Hb_001104_110 |
0.12375774 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Populus euphratica] |
| 3 |
Hb_001268_120 |
0.1297759161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000905_170 |
0.1385776078 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 5 |
Hb_000009_330 |
0.1420613795 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_006538_080 |
0.1450489546 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 7 |
Hb_002631_040 |
0.1457419464 |
- |
- |
hypothetical protein EUGRSUZ_J03178 [Eucalyptus grandis] |
| 8 |
Hb_003044_030 |
0.147077372 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Cicer arietinum] |
| 9 |
Hb_000227_060 |
0.1474229032 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 10 |
Hb_000008_090 |
0.1493653462 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 11 |
Hb_082741_010 |
0.1562737409 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 12 |
Hb_000169_170 |
0.1562850829 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 13 |
Hb_011930_070 |
0.156862014 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 14 |
Hb_000631_120 |
0.1600982074 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 15 |
Hb_049436_010 |
0.1601163866 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |
| 16 |
Hb_008066_070 |
0.1608772805 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 17 |
Hb_003849_240 |
0.1622646621 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000363_400 |
0.1631637546 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 19 |
Hb_007382_120 |
0.1632770269 |
- |
- |
PREDICTED: delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_002344_060 |
0.1633937868 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |