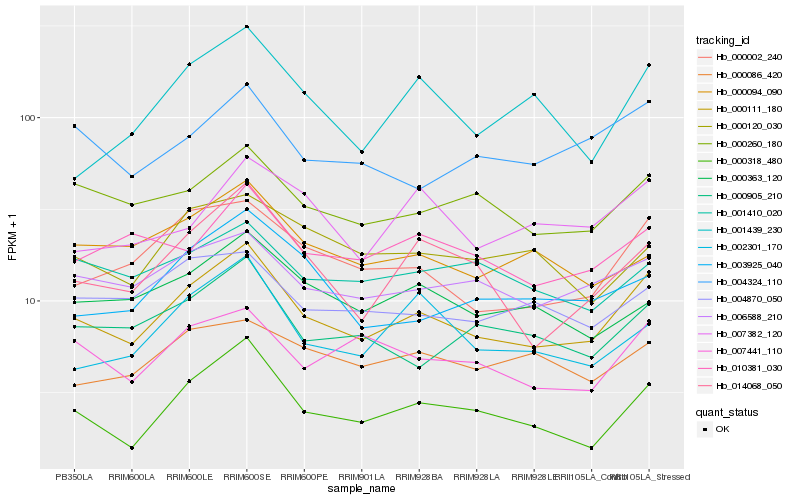
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_180 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 2 |
Hb_000002_240 |
0.0945700227 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 3 |
Hb_000318_480 |
0.0947739233 |
- |
- |
PREDICTED: RING-H2 finger protein ATL65 [Jatropha curcas] |
| 4 |
Hb_006588_210 |
0.1019703211 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 5 |
Hb_014068_050 |
0.1050179178 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 6 |
Hb_000260_180 |
0.1058067207 |
- |
- |
PREDICTED: KH domain-containing protein At2g38610 [Jatropha curcas] |
| 7 |
Hb_007441_110 |
0.1106001864 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At3g58900-like [Jatropha curcas] |
| 8 |
Hb_000905_210 |
0.1123254601 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 9 |
Hb_004324_110 |
0.115183261 |
- |
- |
PREDICTED: abscisic acid receptor PYL8 [Jatropha curcas] |
| 10 |
Hb_010381_030 |
0.1163635874 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 11 |
Hb_000363_120 |
0.1182499059 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 12 |
Hb_004870_050 |
0.1193063914 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 13 |
Hb_002301_170 |
0.1204038792 |
- |
- |
MAC/Perforin domain-containing protein [Theobroma cacao] |
| 14 |
Hb_003925_040 |
0.1208118819 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 15 |
Hb_000094_090 |
0.1223388072 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001410_020 |
0.1230825362 |
- |
- |
cdk8, putative [Ricinus communis] |
| 17 |
Hb_000086_420 |
0.1233883023 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 18 |
Hb_001439_230 |
0.1249736976 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_007382_120 |
0.1257007498 |
- |
- |
PREDICTED: delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000120_030 |
0.1260895171 |
- |
- |
PREDICTED: uncharacterized protein LOC105630385 isoform X1 [Jatropha curcas] |